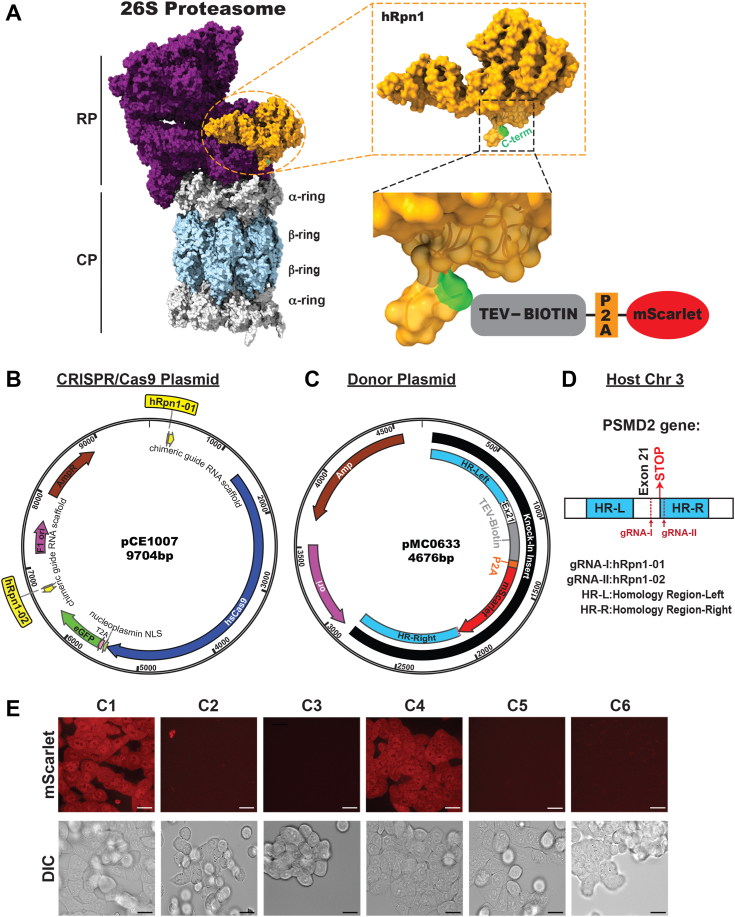
Figure 1.
Overall strategy to generate an engineered cell line with a biotin handle on proteasome subunit hRpn1.A, surface view of a structural state of the human 26S proteasome (left, PDB: 5GJQ) illustrating hRpn1 within the RP (violet) in orange with its C-terminal residue Y908 highlighted in green. The CP α-ring and β-ring subunits are colored gray and light blue, respectively. Enlarged views of hRpn1 are provided to the right of a transparent rendering of the surface diagram with a ribbon diagram for the C-terminal region and in the lower image, including a representation of the knock-in tag containing the TEV-biotin segment (gray box), autocleavable P2A site (orange box), and mScarlet (red oval shaped box). B, CRISPR/Cas9 plasmid map with the two gRNAs (hRpn1-01 and hRpn1-02) represented in yellow. C, donor plasmid map with the knock-in tag consisting of homology region-left (HR-L, blue), exon 21 (E × 21), TEV-biotin (gray), the P2A site (orange), mScarlet (red), and homology region-right (HR-R, blue). D, representation of the region including and neighboring exon 21 of PSMD2 in chromosome 3, highlighting the respective homology regions (HR-L and HR-R) and location of the stop codon and gRNA-binding sites. E, confocal microscopy images captured for six clones (C1–C6) show fluorescent mScarlet signal (top panel) and differential interference contrast (DIC) reference images (bottom panel). The scale bar represents 12 μm. CP, core particle; gRNA, guide RNA; P2A, peptide 2a; PDB, Protein Data Bank; RP, regulatory particle; TEV, tobacco etch virus.