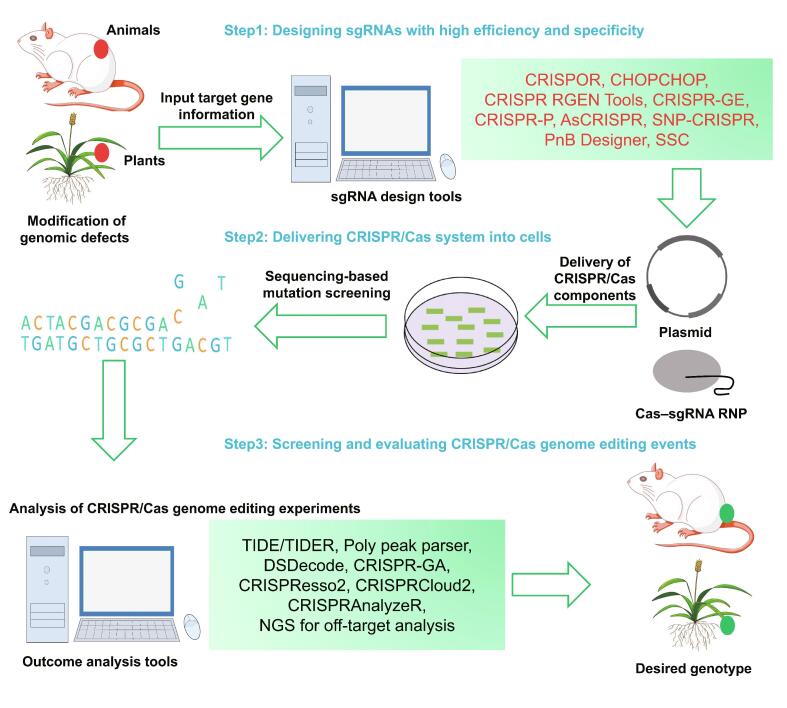
Figure 2.
Example workflow for applying genome editing tools to modify genome sequences
Targeted genome modification has great potentials to be applied in human gene therapy and crop genetic improvement. To proceed a CRISPR/Cas genome editing experiment, the initial step is to design an optimal sgRNA with high efficiency and specificity. On the basis of large-scale empirical data, many algorithm/predictive models have been established and eventually integrated in several web-based applications, such as those shown in upper panel with red color words. Those web-accessible computational tools are designed mainly based on three sets of scoring system, sgRNA efficiency scores, sgRNA specificity scores, and output prediction scores. After performing genome editing experiments, sequencing-based screening will be implemented to evaluate on-target outcomes and off-target effects. To facilitate the efficiency of identifying desired CRISPR/Cas editing events, several web-based resources provide comprehensive computational analysis strategies that meet the needs not only for small-scale genome editing experiments but also for large-scale pooled CRISPR/Cas9 library screening, like those shown in the lower panel with black color words. In addition, many methods and tools have been developed for analyzing outcome off-target effects as listed in Table 2. sgRNA, single guide RNA; NGS, next-generation sequencing; RNP, ribonucleoprotein.