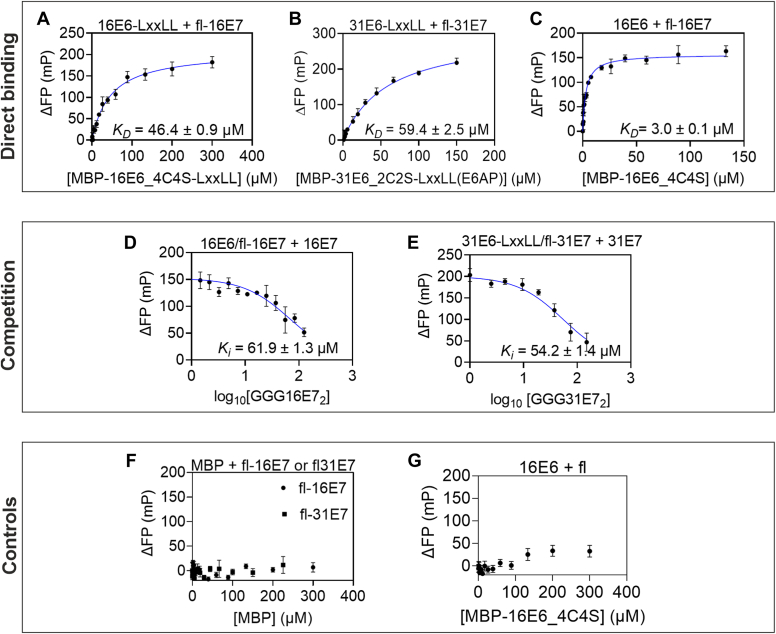
Figure 4.
The binding affinity of the E6–E7 complex.A–C, direct binding curves of purified MBP-E6-LxxLL or MBP-E6 with fl-E7 were monitored in fluorescence polarization by titrating fl-E7 with an increasing amount of E6. All E6 proteins used above consist of C/S mutation. The clear increase in FP indicates a binding event has occurred. The binding affinity of MBP-E6-LxxLL and fl-E7 from HPV16 (A) and HPV31 (B) is similar. MBP-16E6_4C4S (C) shows a 15-fold higher binding affinity than MBP-16E6_4C4S-LxxLL. D and E, the reversibility of the complex formation was monitored with a competitive measurement by titrating the complex with an increasing amount of nonlabeled GGG-E7 dimer. A decrease in the FP signal indicates the reversible complex formation. Concluding from the competition measurement, HPV16 (D) and HPV31 (E) formed E6/E7 complex at a similar binding affinity, and the binding is independent of the LxxLL peptide from E6AP. F, an increasing amount of MBP was titrated against fl-16E7 or fl-31E7. No significant increase in the FP signal indicates that the binding between E6 and E7 is not an artifact of the MBP tag. G, an increasing amount of MBP-16E6_4C4S was titrated against the fluorescein peptide. A slight increase in FP signal at higher concentrations indicates the presence of artifact from fluorescein. All FP signals were subtracted with the FP signal of respective fl-E7 or fl alone and plotted against concentrations of MBP-16E6_4C4S-LxxLL, MBP-31E6_2C2S-LxxLL, MBP-16E6_4C4S, MBP, or nonlabeled GGG-E7 as indicated. The error bar plotted is the standard deviation of the mean from three technical replicates. FP, fluorescence polarization; MBP, maltose-binding protein.