Figure 1.
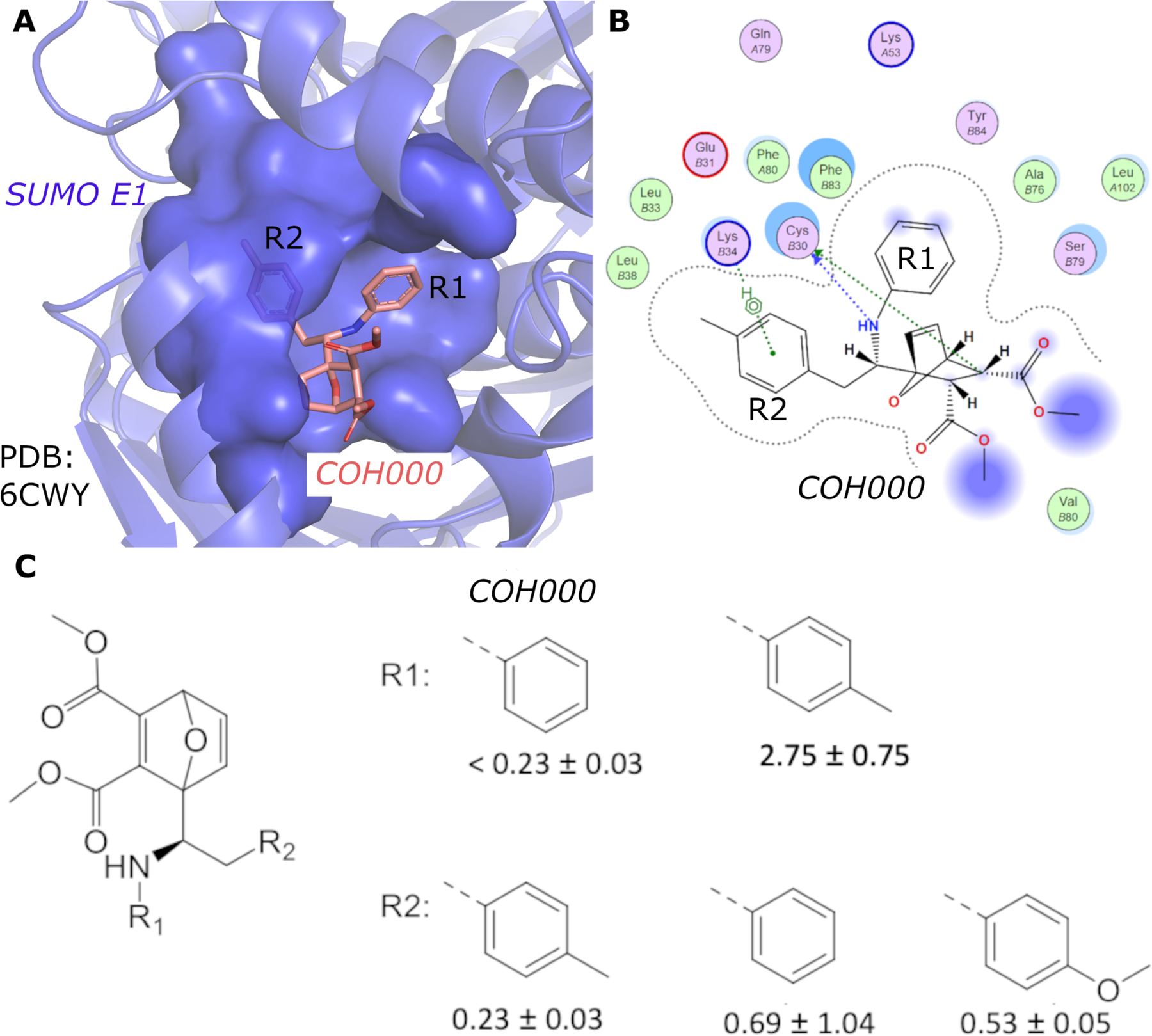
(A) Crystal structure of SUMO E1 (cartoon) in complex with the COH000 allosteric inhibitor (sticks) (PDB: 6CWY). The protein residues of SUMO E1 surrounding COH000 are represented in surface. (B) 2D LigPlot represention of the binding pocket residues of SUMO E1 that interact with COH000. Blue filled circles on COH000 represent solvent accessibility with larger circles representing greater solvent accessibility. Polar and non-polar residues are colored in pink and green circles, respectively. Acidic and basic residues have a red and blue circle border, respectively. (C) Discrepancy between the crystal structure and the structure-activity-relationship of the compounds obtained. Specifically, the R1 group does not tolerate a small change from a benzyl to a methyl benzyl group, although the methyl addition site is partially solvent accessible and has the space to accommodate the addition of a methyl group in the crystal structure. In contrast, variations of the R2 group that is deeply buried in a hydrophobic pocket and is not solvent accessible in the crystal structure tolerates a variety of changes, including removal of the methyl group or replacement of methyl with a methoxy.
