Figure 4 |. Sequencing by synthesis: single-nucleotide addition approaches.
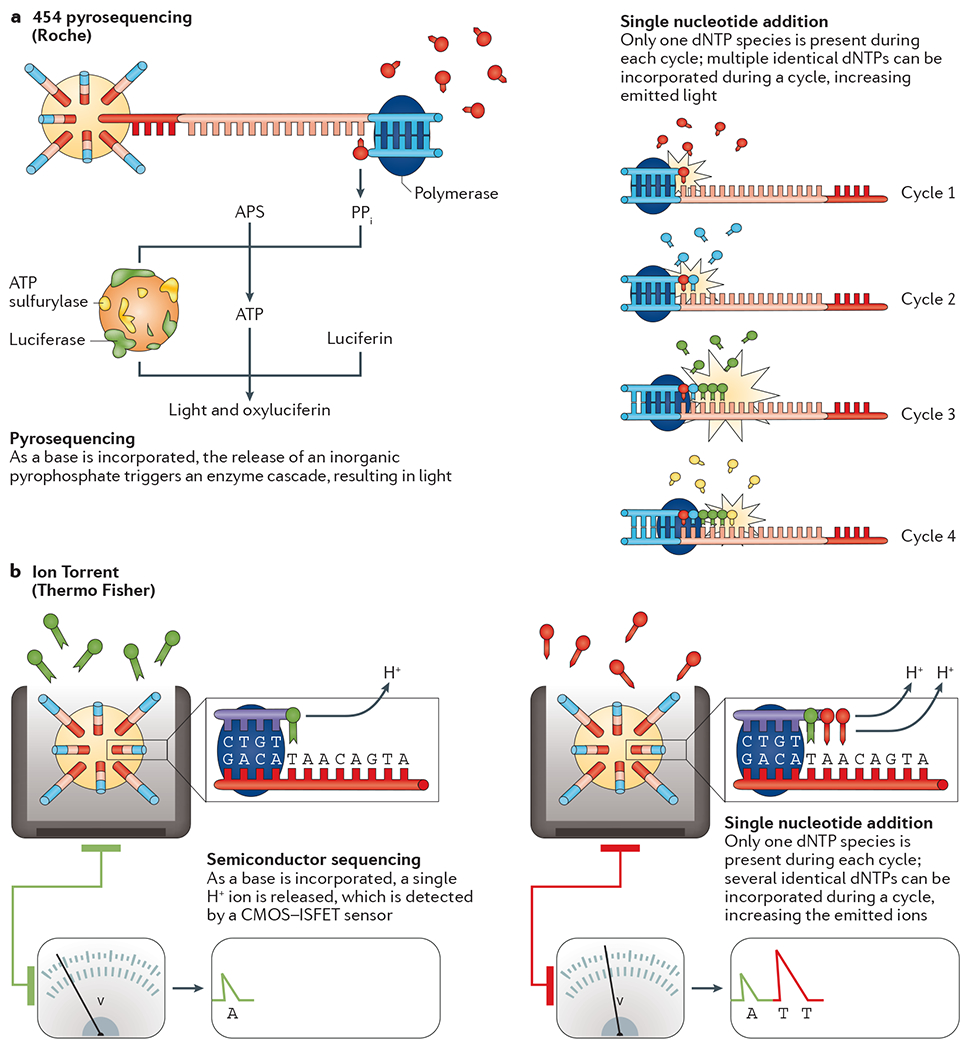
a | 454 pyrosequencing. After bead-based template enrichment, the beads are arrayed onto a microtitre plate along with primers and different beads that contain an enzyme cocktail. During the first cycle, a single nucleotide species is added to the plate and each complementary base is incorporated into a newly synthesized strand by a DNA polymerase. The by-product of this reaction is a pyrophosphate molecule (PPi). The PPi molecule, along with ATP sulfurylase, transforms adenosine 5′ phosphosulfate (APS) into ATP. ATP, in turn, is a cofactor for the conversion of luciferin to oxyluciferin by luciferase, for which the by-product is light. Finally, apyrase is used to degrade any unincorporated bases and the next base is added to the wells. Each burst of light, detected by a charge-coupled device (CCD) camera, can be attributed to the incorporation of one or more bases at a particular bead. b | Ion Torrent. After bead-based template enrichment, beads are carefully arrayed into a microtitre plate where one bead occupies a single reaction well. Nucleotide species are added to the wells one at a time and a standard elongation reaction is performed. As each base is incorporated, a single H+ ion is generated as a by-product. The H+ release results in a 0.02 unit change in pH, detected by an integrated complementary metal-oxide semiconductor (CMOS) and an ion-sensitive field-effect transistor (ISFET) device. After the introduction of a single nucleotide species, the unincorporated bases are washed away and the next is added. Part a is adapted from REF. 18, Nature Publishing Group.
