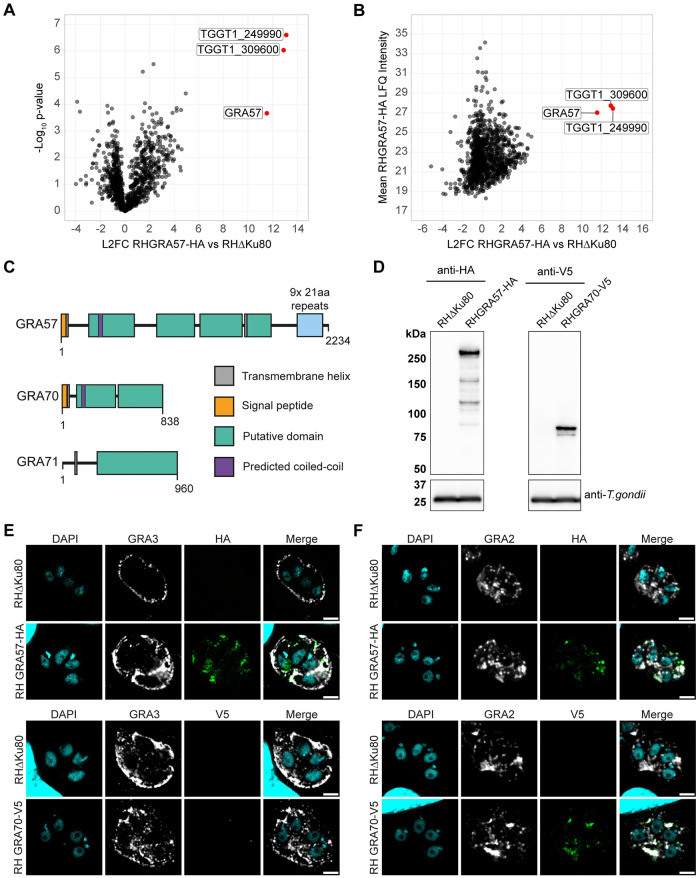
Fig 3. CoIP of GRA57-HA shows it likely forms a complex with TGGT1_249990 and TGGT1_309600 within the PV.
(A, B) Co-immunoprecipitation of endogenously HA-tagged RHGRA57. HFFs were pre-stimulated with 2.5 U/ml IFNγ for 6 h, then infected in triplicate with either RHΔKu80 or RHGRA57-HA for a further 24 h. GRA57 was immunoprecipitated with anti-HA agarose matrix then immunoprecipitate was analysed by mass spectrometry. (A) Enrichment of proteins in RHGRA57-HA vs. RHΔKu80 control samples. (B) Mean LFQ signal intensity detected for proteins in RHGRA57-HA samples. (C) Schematic of putative domains in GRA57, GRA70, and GRA71, to scale. Domains and boundaries were assigned based on regions of predicted secondary structure using PsiPred. Transmembrane helices are annotated where predicted by at least 2 programmes (MEMSAT-SVM, DeepTMHMM, and CCTOP). Regions of predicted coiled-coil were determined using Waggawagga. (D) Western blot protein expression analysis of endogenously tagged lines. (E) Co-localisation of endogenously c-terminally tagged GRA57 and GRA70 with the PVM marker GRA3, which is exposed to the host cell cytosol. Samples were permeabilised with 0.1% saponin. Scale bar = 3 μm. (F) Co-localisation of endogenously c-terminally tagged GRA57 and GRA70 with the IVN marker GRA2. Samples were permeabilised with 0.2% Triton-X100. Scale bar = 3 μm. Source data for A and B can be found in S4 Data. HFF, human foreskin fibroblast; IVN, intravacuolar network; LFQ, label-free quantification; PV, parasitophorous vacuole; PVM, parasitophorous vacuole membrane.