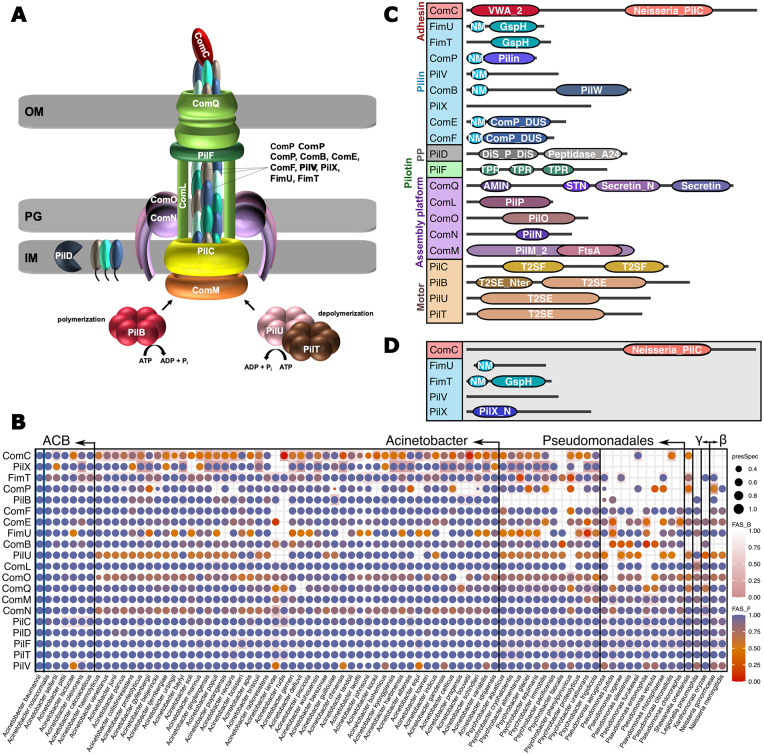
Fig 1. Characterization of Acinetobacter type IVa pilus components.
(A) Model of the type IVa pilus (T4aP) in A. baylyi. OM–Outer membrane; IM–Inner membrane; PG–peptidoglycan. The individual components are listed in Table 1. (B) Phylogenetic profiles of the Ab ATCC 19606 T T4aP components across 884 bacterial isolates representing 83 named species. Taxa are summarized on the species level. A dot indicates the presence of an ortholog in the respective species, and the dot size represents the fraction of the subsumed isolates an ortholog was identified in. The color encodes the median feature architecture similarity (FAS score) between the protein in Ab ATCC 19606 T and its orthologs within a species. The dot color gradient (FAS_F; blue to orange) captures architecture differences using the feature architecture of the Ab ATCC 19606T protein as reference. The score decreases if features seen in the Ab ATCC 19606T protein are missing in the respective orthologs. The cell color gradient (FAS_B; white to pink) captures architecture differences using the feature architecture of the ortholog as reference. The score decreases if features seen in the ortholog are missing in the Ab ATCC 19606T protein. Taxa are ordered according to increasing phylogenetic distance to A. baumannii using the information provided in [21] and [100]. ‘γ‘ and ‘β‘ represent γ- and β-proteobacteria, respectively. The full data is available as S1 Data. (C) Pfam domain architectures of the T4aP components in Ab ATCC 19606 T. Protein lengths are not drawn to scale. Pfam accessions and domain descriptions are available in S1 Table. PP–Prepilin peptidase; NM–N_Methyl. (D) Most common alternative domain architectures for ComC, FimU and FimT represented in (C). Architectures represent ComC, FimU, and PilV in A. baumannii 1297, FimT in A. baylyi, and PilX in A. baumannii AYE.