Fig. 1.
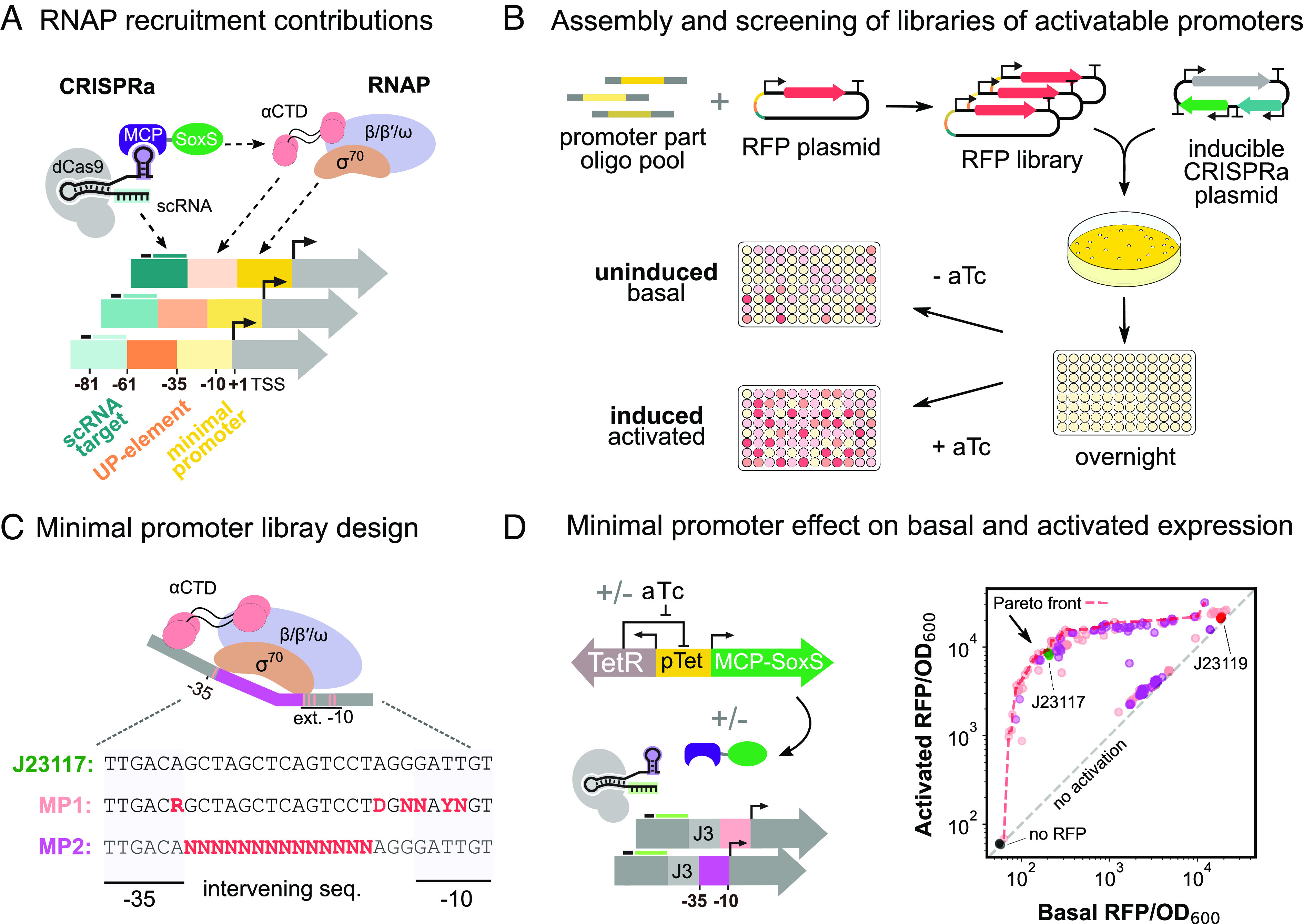
Functional interrogation of promoter regions with CRISPRa. (A) Schematic of RNAP interactions with the CRISPRa complex and target promoter. σ70 affinity for the minimal promoter and α-CTD affinity for the UP element determines RNAP recruitment to a promoter. When CRISPRa is targeted to a promoter with a complementary scRNA target site, the RNAP α-CTD domain is recruited by the SoxS transcriptional activator. RNAP-promoter and CRISPRa-promoter interactions can be modulated by modifying the DNA sequence of the different promoter regions. (B) Workflow for the assembly and characterization of libraries of activatable promoters. A library of RFP genes with varying promoters is generated through PCR (Methods S9). The library is then cotransformed into E. coli with an aTc-inducible CRISPRa plasmid. Colonies are then seeded overnight, and subsequently diluted into media with appropriate concentrations of aTc. For each promoter variant in the libraries, basal and activated RFP levels were measured with 0 nM and 200 nM aTc, respectively (Methods S10. (C) Schematic of RNAP interaction with the minimal promoter and library design. σ70 recognizes specific positions in the extended −10 and −35 regions of the minimal promoter, which informed the design of the library MP1. σ70 binding is also influenced by the GC content, the length, and the −15TGn−1 motif of the intervening sequence, which informed the design of library MP2 (Methods S11). (D) Minimal promoter effect on expression levels. Left: Inducible CRISPRa system and minimal promoter libraries of the J3 synthetic promoter. MCP-SoxS is expressed from the aTc-inducible pTet promoter. dCas9 and J306 scRNA are constitutively expressed. Right: Activated and basal RFP/OD600 for the two minimal promoter libraries (nMP1 = 89, nMP2 = 84). The red dash line defines the Pareto front containing the best-performing promoter variants (SI Appendix, Methods S3), for which no further improvements in basal or activated levels can be achieved without compromising the other. The gray dash line defines promoter variants with equal activated and basal expression levels, indicating they are not activated by CRISPRa. The J23117 minimal promoter (green, triplicates) is included as a standard reference for CRISPRa efficiency. The J23119 minimal promoter (red, triplicates) is an example of a nonactivatable promoter due to high basal expression levels. A plasmid without RFP (black, triplicates) indicates the background fluorescence of the system.
