Fig. 4.
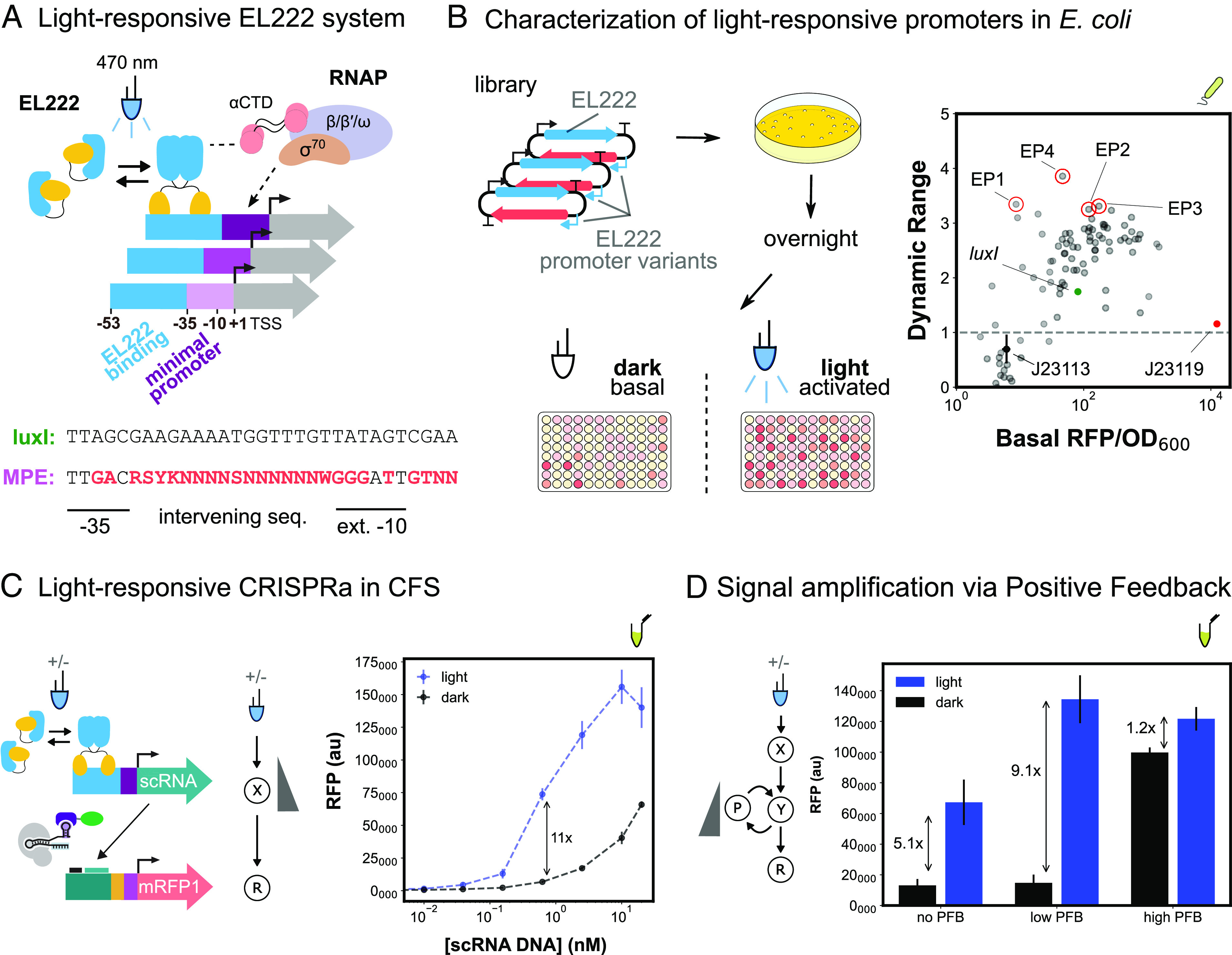
Developing activatable promoters for blue light–responsive CRISPRa/i circuits. (A) Schematic of EL222 light-responsive promoter system and library design. EL222 transcription factor dimerizes in response to 470-nm light and binds a specific sequence upstream of the minimal promoter. EL222 then recruits RNAP through interactions with the α-CTD domain. The minimal promoter library design is based on the original luxI promoter and previous minimal promoter libraries (Methods S11). (B) Characterization of light-responsive promoters in E. coli. Left: Blue-light promoter screening (Methods S13). EL222 protein and promoter library are expressed from a single plasmid. Assembly and screening are carried out as previously described. Basal and activated expression levels are measured from cultures not exposed or continuously exposed to blue light, respectively. Right: Basal expression and dynamic range of blue-light promoter variants (nMP3 = 96). The gray dash line defines promoter variants with equal activated and basal expression levels, indicating they are not activated by EL222. The J23119 minimal promoter (red) and J23113 (black) are examples of nonactivatable promoters. Variants with improved performance (red circles) compared to the original luxI promoter (green) were selected for use in CFS. (C) Light-responsive CRISPRa in CFS. Left: EL222 scRNA expression from an engineered blue-light promoter and downstream CRISPRa. Reactions contain 8 nM and 10 nM of EL222 and RFP plasmids respectively. Right: Titration of blue light–inducible scRNA plasmid concentration to maximize the fold change between blue light–dependent CRISPRa (blue) and CRISPRa due to scRNA leak in the dark (black). (D) Improvement of blue-light CRISPRa dynamic range through the construction of a PFB circuit. Left: Blue light–responsive CRISPRa cascade with PFB. PFB is achieved by including a downstream node that expresses a scRNA targeting an upstream node Reactions contain 15 nM and 10 nM of EL222 and RFP plasmids respectively. Right: Blue light–dependent CRISPRa (blue) and CRISPRa due to scRNA leak in the dark (black). The amount of PFB was tuned by adjusting the concentration of the PFB node. “No”, “Low”, and “High” PFB concentrations correspond to 0, 3 pM, and 2 nM, respectively. For all panels, values represent the mean ± SD of three technical replicates.
