Fig. 5.
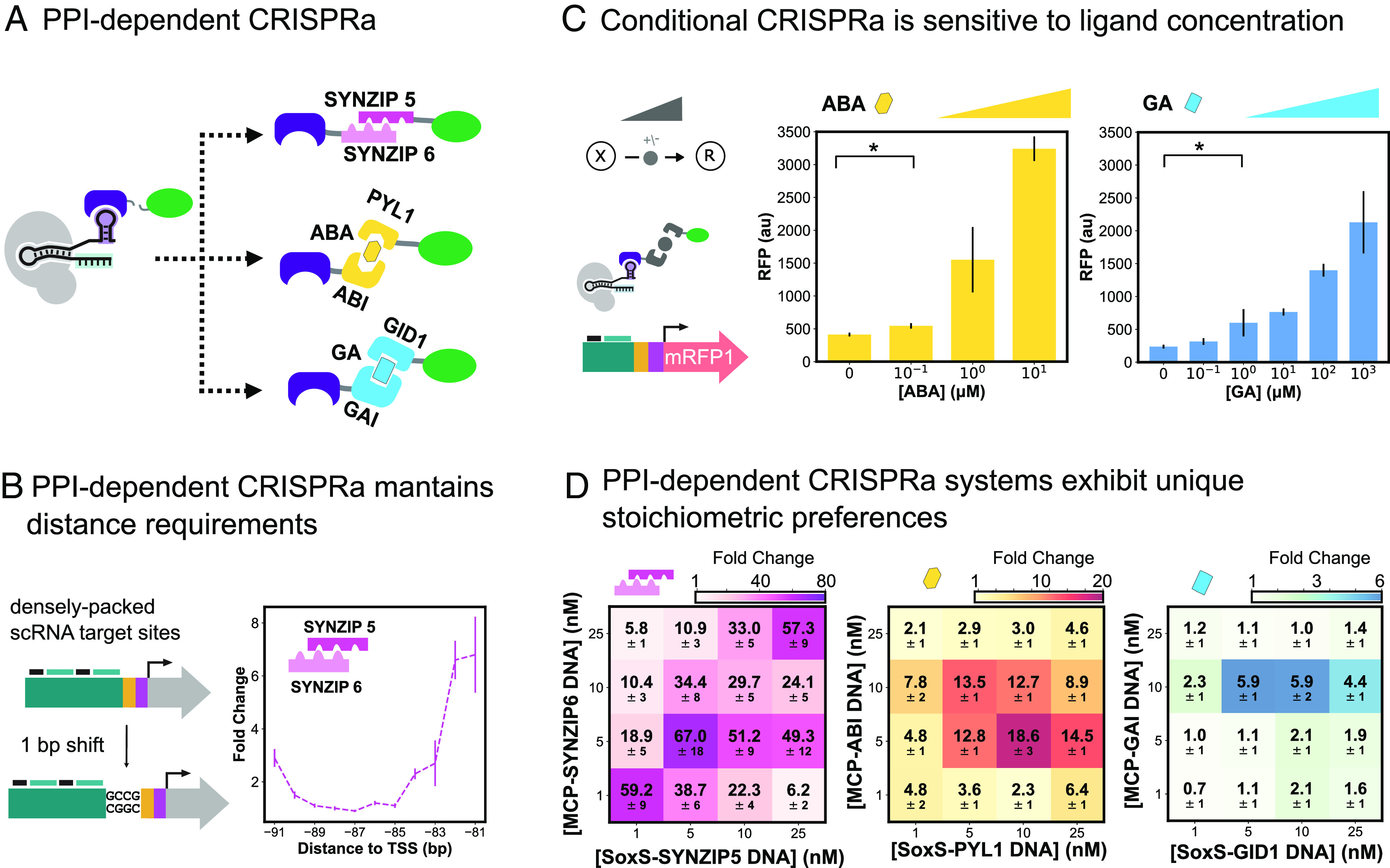
Engineered activatable promoters enable PPI-dependent conditional CRISPRa. (A) Schematic of different PPI-dependent CRISPRa systems. MCP-SoxS fusion is split and the two proteins are instead fused to one end of a heterodimerization domain. The heterodimerization domains used to build PPI-dependent CRISPRa systems are the SYNZIP5/SYNZIP6 pair, the ABA-responsive ABI/PYL1 domain, and the gibberellic acid (GA)-responsive GAI/GID1 domain. (B) Distance requirements of PPI-dependent CRISPRa. Left: Engineered promoter containing densely packed scRNA target sites and single base pair 5′ additions allows for CRISPRa targeting between −81 and −111 bp from the TSS. Right: Testing SYNZIP-CRISPRa between −81 and −91 bp from the TSS. SYNZIP-CRISPRa components are expressed at 5 nM. Fold change is calculated relative to an off-target scRNA for each promoter variant. (C) Tuning conditional CRISPRa response through titration of small molecule concentration. For ABA- and GA- CRISPRa, the corresponding small molecule was titrated between 0 and 10 or 0 and 103 μM, respectively to find the optimal concentration. ABA- and GA-CRISPRa components are expressed at 10 nM. (D) Improving PPI-dependent and conditional CRISPRa response by optimizing component stoichiometries. The concentration of the plasmids expressing the MCP and SoxS components for each dimerization system was varied 1 to 25 nM and tested combinatorially to find the best ratio of the two heterodimers. ABA is added at 10 μM and GA is added at 103 μM. Fold change is given relative to a reaction with no MCP and SoxS plasmids added.
