Fig. 6.
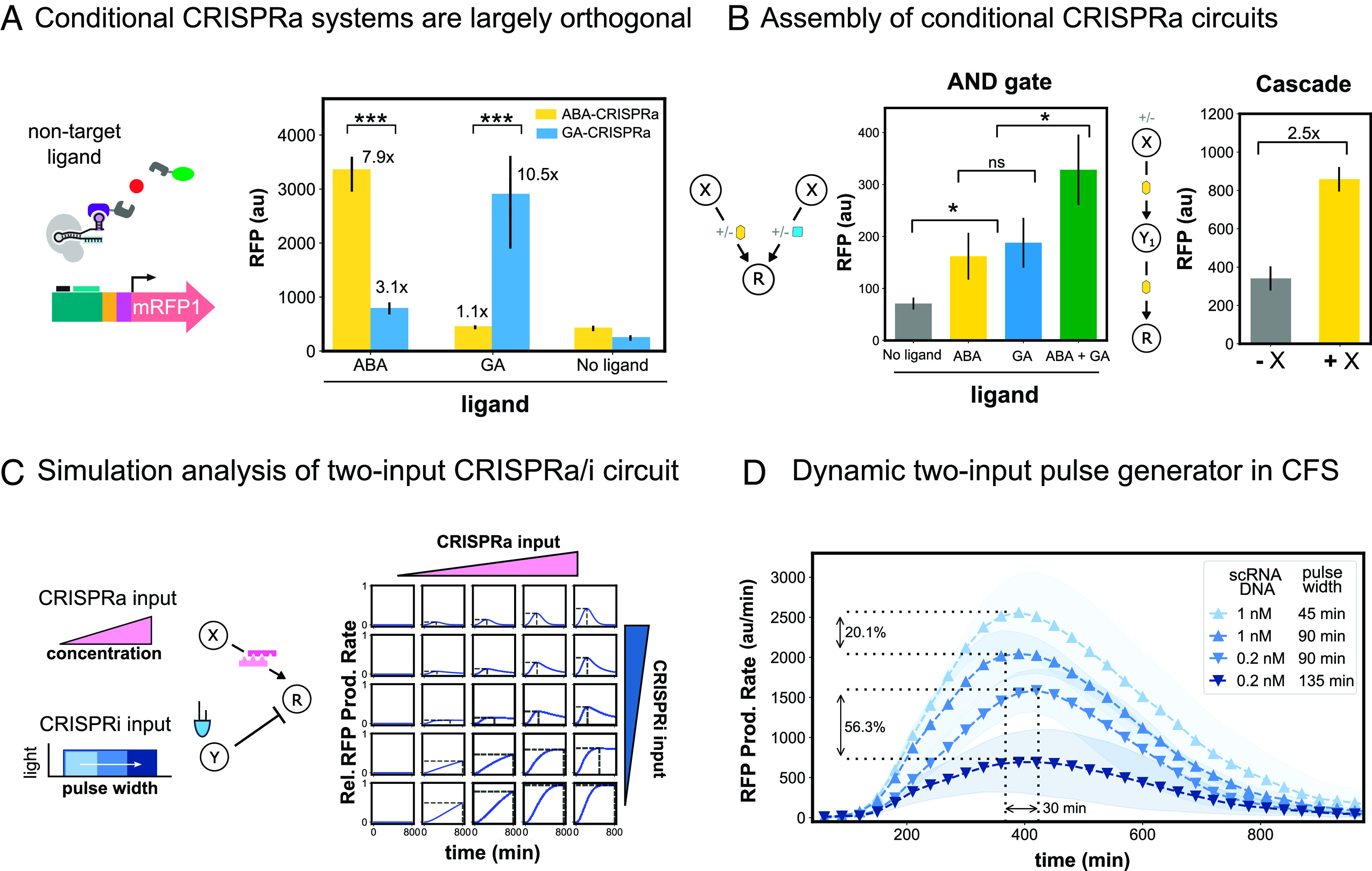
Engineering multi-input CRISPRa/i circuits. (A) Conditional CRISPRa response to noncognate ligands. The orthogonality of the small molecule-responsive conditional CRISPRa systems was tested by adding either the corresponding or noncorresponding small molecule to cell-free reactions containing the components for ABA- or GA-CRISPRa. All components are added at their respective optimal screened concentrations. ABA is added at 10 μM and GA is added at 103 μM. (B) Assembly of conditional CRISPRa circuits. For both circuits, all components are added at their respective optimal screened concentrations. ABA is added at 10 μM and GA is added at 103 μM. Left: AND-like behavior was constructed by adding the components for both ABA- and GA- CRISPRa in a cell-free reaction. Right: The CRISPRa cascade was assembled by using ABA-CRISPRa to activate expression of both the first and second nodes in an activation cascade. The first node was added at either 0.05 or 0 nM, and the internal and output nodes were added at 10 nM. (C) Simulation analysis of a two-input CRISPRa/i circuit using SYNZIP5/SYNZIP6 heterodimerization mediated-CRISPRa and blue-light CRISPRi (CFS Blue-light CRISPRa/imodeling). (D) SYNZIP-CRISPRa and blue-light CRISPRi were integrated to construct a tunable pulse generator. The amount of CRISPRa was tuned by adding either 0.2 nM or 1 nM of constitutively expressed scRNA plasmid to the CFS reaction. The sgRNA targeting RFP for CRISPRi was driven from the blue light–responsive engineered EL222 promoter. The amount of CRISPRi was tuned by adjusting the time of blue-light exposure between 45 and 135 min. RFP production rates (SI Appendix, Methods S3) are plotted as a function of CRISPRa and CRISPRi inputs. For all panels, values represent the mean ± SD of three technical replicates.
