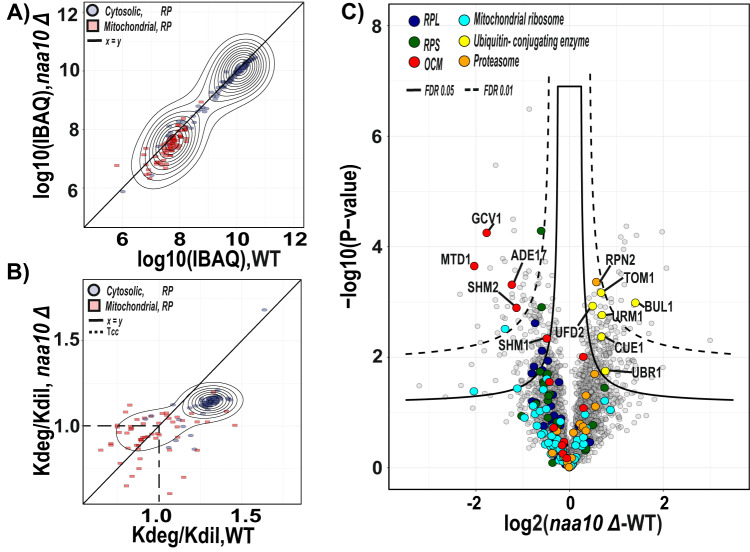
Fig. 5. NatA-defective yeast cells compensate for fast ribosomal turnover rate by adjusting protein synthesis.
A Scatterplot comparing log10 of the intensity-based absolute quantification (IBAQ) of cytosolic and mitochondrial ribosomal proteins between WT and naa10Δ strains. n = 3 replicate yeast cultures per condition. B Same as (A) but comparing the normalized turnover rate between conditions. n = 2 replicate cultures. Diagonal line in (A, B) correspond to the identity function. Dashed lines indicate the cell cycle time (Tcc). C Comparison of the WT and naa10Δ strains as a volcano plot to identify newly synthesized protein abundance significant changes. Significant regulated proteins at 1% and 5% false discovery rate (FDR) are delimited by dashed and solid lines, respectively (FDR controlled, two-sided t test, randomizations = 250, s0 = 0.1). Source data are provided as a Source Data file (A–C).