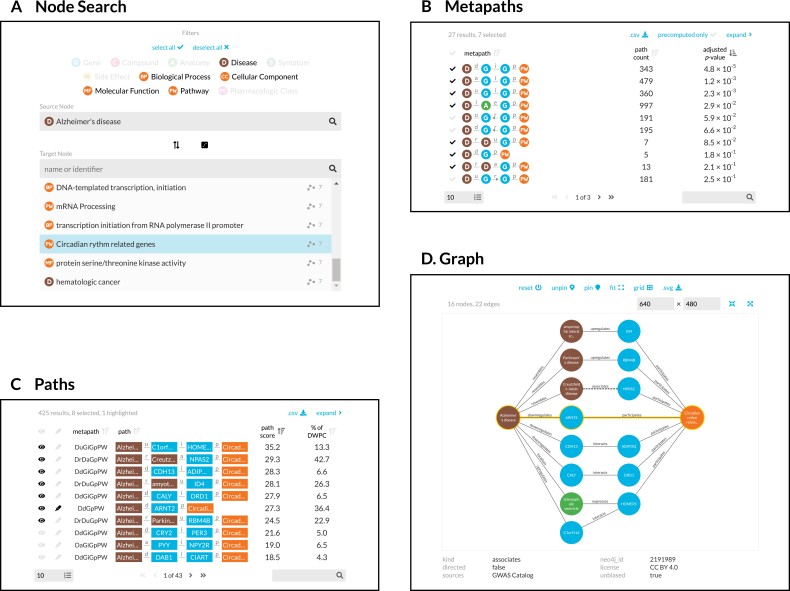
Figure 3:
Using the connectivity search webapp to explore the pathophysiology of Alzheimer's disease. This figure shows an example user workflow for https://het.io/search/. (A) The user selects 2 nodes. Here, the user is interested in Alzheimer's disease, so selects this as the source node. The user limits the target node search to metanodes relating to gene function. The target node search box suggests nodes, sorted by the number of significant metapaths. When the user types in the target node box, the matches reorder based on search word similarity. Here, the user becomes interested in how the circadian rhythm might relate to Alzheimer's disease. (B) The webapp returns metapaths between Alzheimer's disease and the circadian rhythm pathway. The user unchecks “precomputed only” to compute results for all metapaths with length ≤3, not just those that surpass the database inclusion threshold. The user sorts by adjusted P value and selects 7 of the top 10 metapaths. (C) Paths for the selected metapaths are ordered by their path score (limited to 100 paths for each metapath). The user selects 8 paths (1 from a subsequent page of results) to show in the graph visualization and highlights a single path involving ARNT2 for emphasis. (D) A subgraph displays the previously selected paths. The user improves on the automated layout by repositioning nodes. Clicking an edge displays its properties, informing the user that an association between Creutzfeldt–Jakob disease and NPAS2 was detected by genome-wide association study.