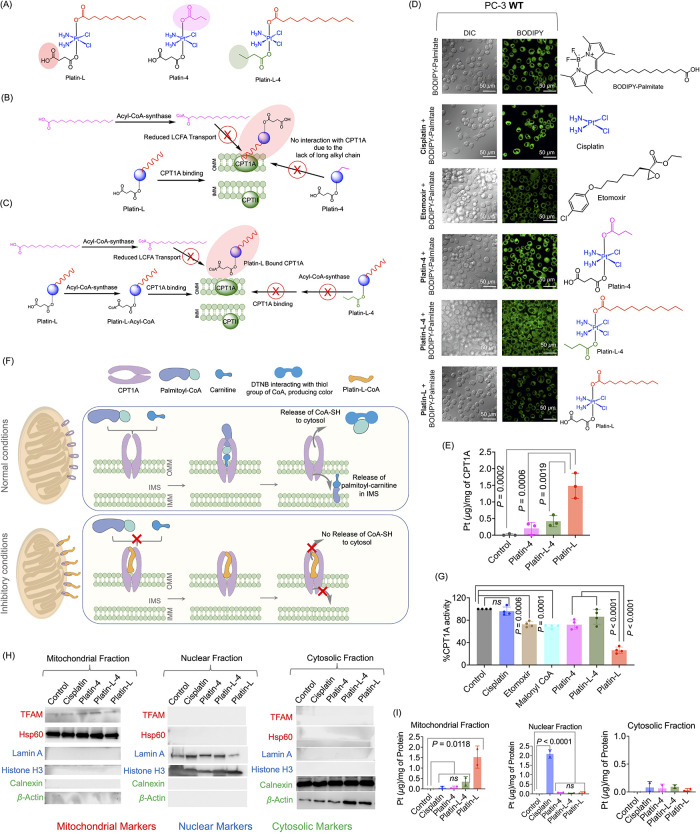
Figure 3.
Effects of axial ligands on CPT1A activity. (A) Structures of Platinum(IV) prodrugs with variation at the axial ligands for understanding the role of these ligands in CPT1A binding. Schematic representation of (B) Platin-L binding to CPT1A due to the long-chain fatty acid, compared to Platin-4 which does not bind due to the short-chain fatty acid and (C) Platin-L binding to CPT1A due to its ability to interact with acyl-CoA synthase to become Platin-L-Acyl-CoA and interact with CPT1A compared to Platin-L-4, which does not have the ability to convert to an acyl-CoA form. (D) Incorporation of fluorescent, long-chain fatty acid BODIPY-palmitate into PC3 cells after treatment with various compounds using live cell imaging. (E) Immunoprecipitation (IP) of CPT1A to quantify the amount of Platinum present on CPT1A after treatment of PC3 cells with Platin-4, Platin-L-4, or Platin-L. The data presented here is an average with the standard deviation from three independent biological replicates (N = 3). (F) Schematic representation of the CPT1A activity assay. Top: normal conditions for CPT1A function, resulting in the release of CoA-SH into the cytosol. Bottom: inhibitory conditions, where CPT1A is inhibited and CoA-SH is not released into the cytosol. The amount of CoA-SH released can be directly correlated to the activity of CPT1A. (G) Relative activity of CPT1A after treatment with test articles at 10 μM. The data presented here is an average with the standard deviation from four independent biological replicates (N = 4), each with three technical replicates (n = 3). Ordinary one-way ANOVA was used to calculate the significance. (H) Western blot analyses of the subcellular fractions used for Pt quantification to ensure purity. (I) Platinum content in subcellular fractions by ICP-MS. The data presented is the mean with the standard deviation from two independent biological replicates. Ordinary one-way ANOVA with multiple comparisons was used to calculate the significance.