Figure 6. In vivo CRISPR screening identifies metabolism pathways regulating the anti-tumor immunity.
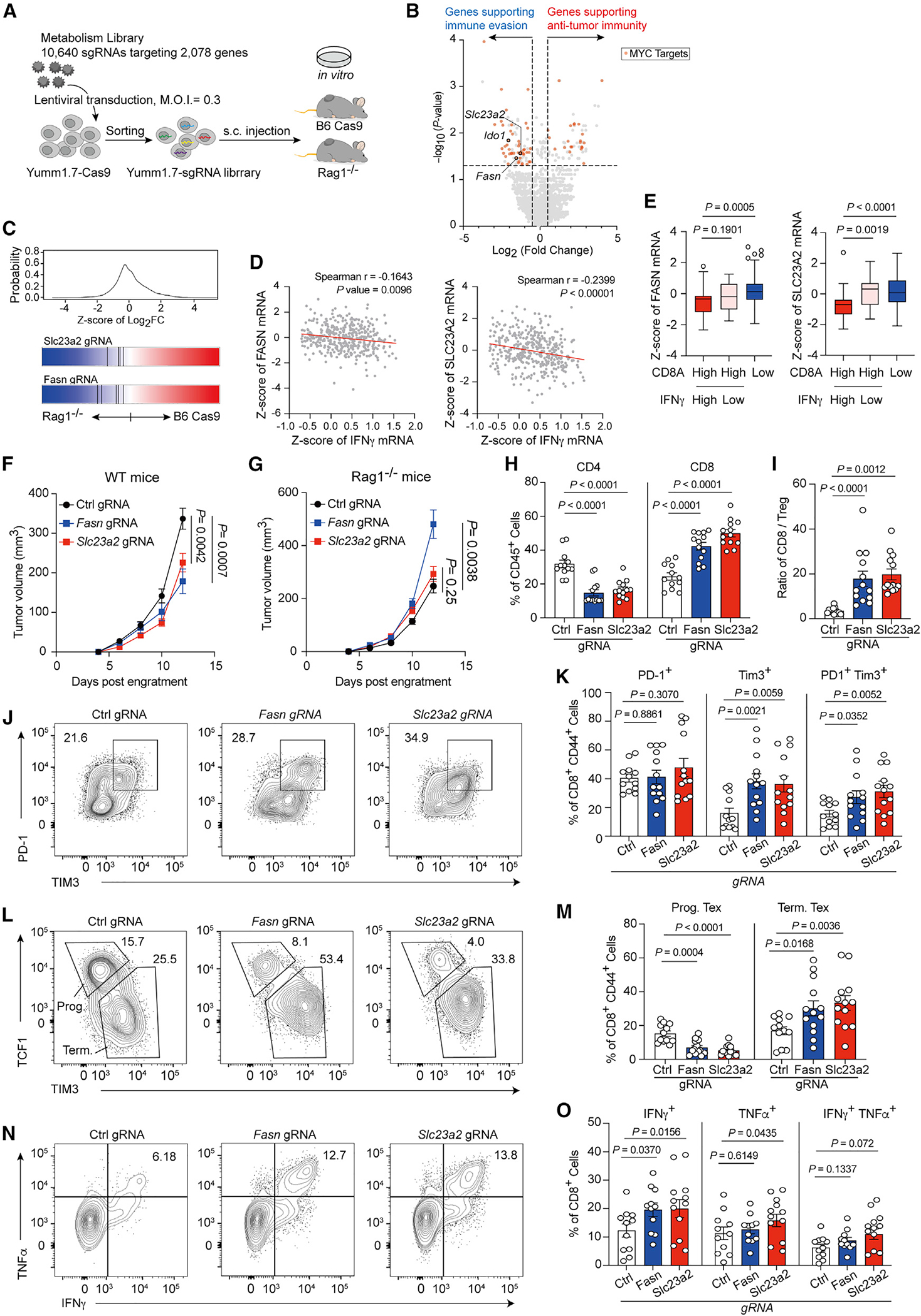
(A) Diagram of in vivo screening system for metabolism-guided immunoediting.
(B) Hits from screen for regulating tumor immunity. The x axis shows the log2 (fold change) comparing gRNA levels of tumors from WT (B6 cas9) with those from Rag1−/− mice, and the y axis denotes the −log10 of p value for differential expression obtained by MAGeCK. (Red symbol represents c-Myc target genes; Ido1, Fasn, and Slc23a2 are highlighted as open circle.)
(C) Frequency histograms of enrichment (Z score) for sgRNAs targeting indicated genes.
(D) Spearman correlation of Fasn (left) or Slc23a2 (right) expression (Z score) with IFNγ (n = 401 biologically independent melanoma tumor samples from TCGA cohort).
(E) Boxplot with Tukey whiskers showing varied Fasn (left) or Slc23a2 (right) expression (Z score) in CD8Ahigh and CD8Alow melanoma patients from TCGA cohort (n = 272).
(F and G) Tumor volume for Yumm1.7 tumors with control gRNA or gene deletions as indicated in WT (F) and Rag1−/− mice (G).
(H) Population of CD4+ (right) and CD8+ (left) T cells in Fasn-null, Slc23a2-null, or control tumors.
(I) Ratio of CD8+ T cells/FoxP3+ Treg in Fasn-null, slc23a2-null, or control tumors.
(J–M) Representative plots and percentages of the PD-1+ Tim3+ T cells (J and K) and the TCF1+ Tim3+ T cells (L and M) among total tumor-infiltrating CD44+ CD8+ T cells from the indicated mice.
(N and O) Representative plots (N) and percentages (O) of the IFNγ and TNF-α-producing tumor-infiltrating T cells from the indicated tumor samples after ex vivo restimulation with CD3/CD28.
Data are the cumulative results from at least two independent experiments (F–O). Each symbol represents one individual. Data are means ± SEM and analyzed by two-tailed, unpaired Student’s t test.
