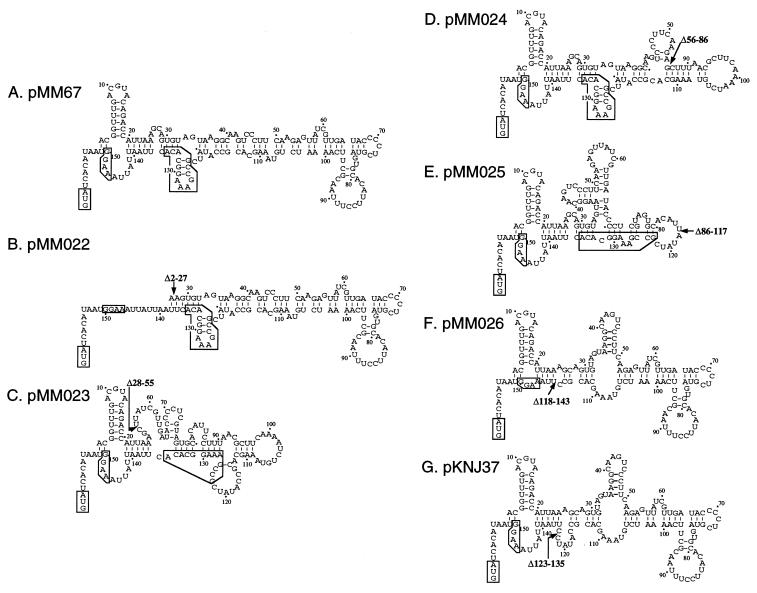
FIG. 6.
Comparison of the secondary structures of the 5′-UTRs for the deletion constructs. Secondary structures of the 5′-UTR for each deletion construct were predicted with a nucleotide sequence analysis program (DNASIS-Mac; Hitachi Software Engineering Co. Ltd.) based on the method of Zuker and Stieger (36). Nucleotides are numbered as the position in the cspA mRNA starting from the transcription initiation site as +1. The position of the deletion in each mutant is shown by an arrow with the nucleotide numbers of the deleted region. The highly conserved 13-base sequences upstream of the SD sequence designated the UBs are boxed. The initiation codon and the SD sequence are also boxed.