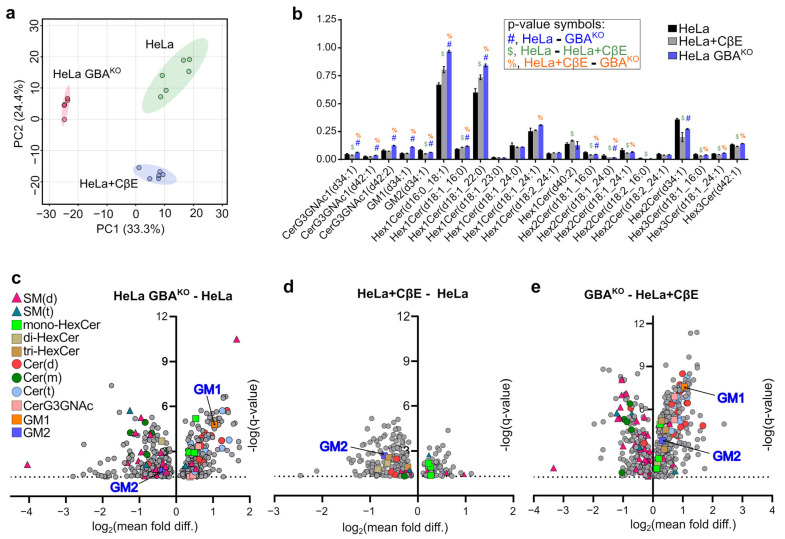
Figure 8.
HeLa, HeLa GBAKO, and HeLa cells treated with CβE have distinct lipidome, GSL, and sphingolipidome profiles. (a) Untargeted lipidomics data were collected on HeLa, HeLa GBAKO, and HeLa cells treated with 35 µM CβE for 2 weeks (HeLa + CβE). HeLa and HeLa + CβE had n = 6 technical replicates, while HeLa GBAKO had n = 5. A total of 845 lipid species were identified in data and were used to conduct a PCA analysis using MetaboAnalyst 5.0. A post hoc Tukey test with false discovery rate correction was used to compare the normalized annotated lipids every two groups, and the adjusted p-values of 0.05 and 0.1 were employed for significant differences. Hierarchical clusters were generated with R (version 4.2). (b) ANOVA corrected for multiple comparisons using FDR at Q = 5% (Prism) was used to identify statistically significant GSLs in untargeted data. Symbols used to indicate significant discoveries in pairwise comparisons are shown in the inset. (c) Volcano plots of indicated comparisons showing significant discoveries following ANOVA corrected for multiple comparisons using FDR at Q = 5%. Plots have Y-axes of −log10 (q-value), and X-axes are log2 of the ratio of means for the indicated comparisons: (c) HeLa GBAKO/HeLa; (d) HeLa + CβE/HeLa; (e) GBAKO/HeLa + CβE.