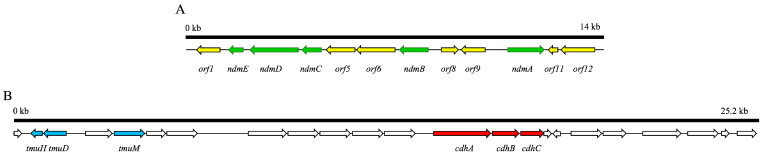
Figure 3.
Characteristics of structures and sequences from the enzymes in the bacterial degradation pathway of caffeine. (A) Sequence arrangement of N-demethylase-related genes in the genome of Pseudomonas sp. NCIM 5235 [58]. Arrows suggest the position and orientation of each orf. Green arrows suggest the genes directly involved in caffeine metabolism, and yellow arrows indicate genes involved in the metabolism of by-products and other unknown functions. (B) Sequence arrangement of genes involved in the C-8 pathway in the genome of Pseudomonas sp. CBB1 [50]. Arrows suggest the position and orientation of each orf. Genes required for caffeine oxidation are represented by blue arrows, while those required for trimethyluric acid oxidation are represented by red arrows.