Table 2.
Studied enzymes with detectable RNA binding.
| Protein | Description of Enriched RNA Features | Predicted RNA Secondary Structure a |
|---|---|---|
| pyruvate kinase (PykF) |
various mRNAs/asRNAs with no apparent communal attribute | - |
| phosphoglycerate kinase (Pgk) | poorly defined hairpins, showing A-rich loops of variable length as common feature |
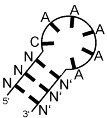
|
| glyceraldehyde-3-phosphate dehydrogenase (GapA) | poorly defined, AU-rich sequence stretches |
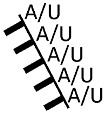
|
| thymidylate synthase (ThyA) |
hairpin with strictly conserved sequence GC(AGA)GU (brackets indicating triloop) |
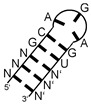
|
| aconitase (AcnB) |
poorly defined stem-loops with 3′-adjacent U-rich stretches |
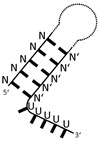
|
| glutamate-5-kinase (ProB) |
small number of specific fragments (two mRNAs and one asRNA) | - |
| quinone oxidoreductase (QorA) | few specific mRNAs/asRNAs with a potential motif (see Figure 8A). top-scoring: yffO mRNA fragment. |
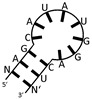
|
| positive control MS2 phage coat protein |
bacteriophage operator hairpin-like motif NANN(ANCA)N′N′N′ (N and N′ indicate complementary bases, brackets indicate loop) |
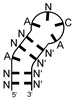
|
a manually curated structures based on different predictions, see Section 4.
