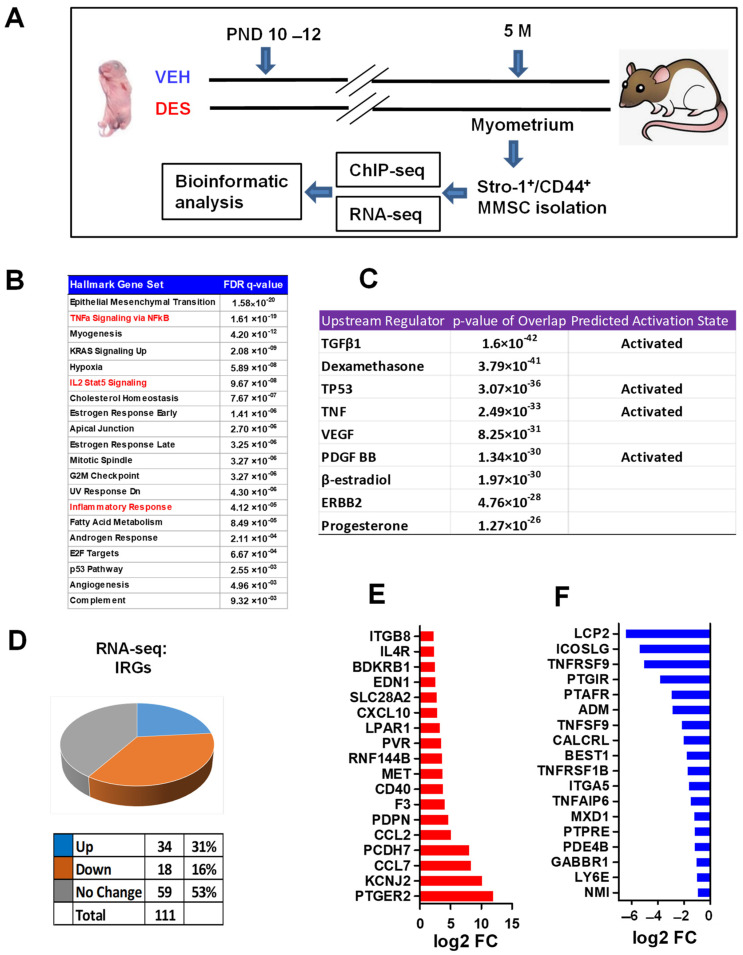
Figure 1.
Developmental DES-exposure-activated inflammation pathway in MMSCs. (A) Experimental paradigm. Eker rat pups were exposed to VEH and DES at postnatal days 10–12, respectively. The pups were euthanized at five months of age, representing the early adult stage. Myometrial tissues were isolated from the animals and subjected to MMSC isolation using Stro-1/CD44 surface markers. Myometria from five animals were pooled for each treatment. Multi-omics analyses, including RNA-seq and ChIP-seq, were performed to determine the transcriptome and histone modification alterations, respectively. (B) Hallmark gene set analysis between DES-MMSCs and VEH-MMSCs. Inflammation-related pathways are highlighted in red. (C) Upstream regulator and predicted activation analysis via Ingenuity Pathway Analysis (IPA). (D) Pie chart showing the percentage of IRGs that exhibited changes in RNA expression between DES-MMSCs and VEH-MMSCs, as measured via RNA-seq; the cutoff value is 2-fold with an FDR < 0.05. (E) List the top 18 IRGs showing differential upregulation in DES- vs. VEH-MMSCs. (F) List the top 18 IRGs showing downregulation in DES- vs. VEH-MMSCs.