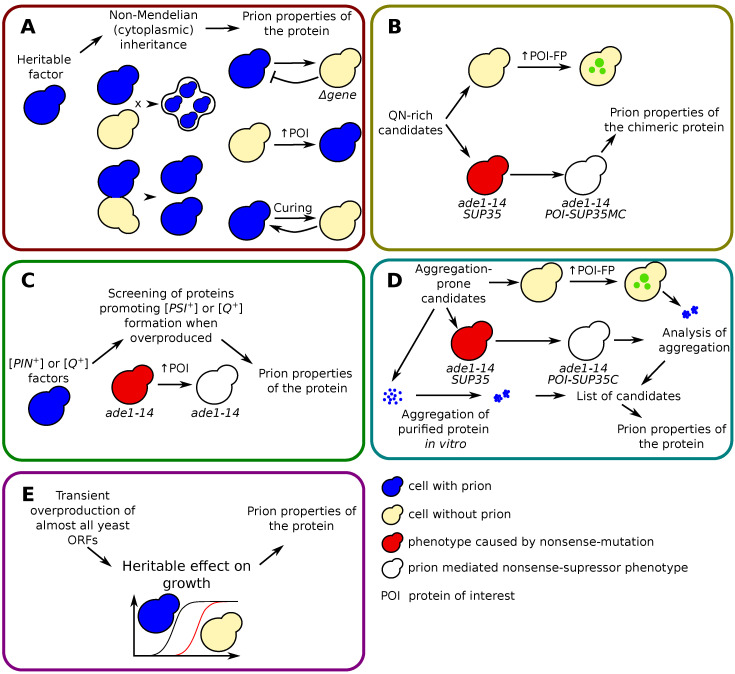
Figure 1.
Strategies for identification of prions or prion-like proteins. (A) The description of non-Mendelian inheritable factors followed by identification of protein required for its propagation. (B) Investigation of prion properties of Q/N-rich proteins. Corresponding candidates are revealed with the bioinformatic analysis of yeast proteome. Aggregation of the protein fused with fluorescent protein is monitored with microscopy. The fusion of protein of interest with functional domain of Sup35 allows verify aggregation on the phenotypic level. Inactivation of the POI-Sup35MC into aggregates leads to the nonsense-suppressor phenotype which can be easily monitored by in cells with nonsense-mutations (for instance, ade1-14). (C) Screening for proteins, which can trigger formation of other prions. Appearance of [PSI] and [Q] factors, leading to the nonsense-suppressor phenotype, requires another prions. Identification of corresponding proteins was carried out with large scale screening when different genes were overexpressed. (D) Analysis of prion properties of aggregation-prone proteins by complex approach: analysis of aggregation of overproduced protein fragments fused with GFP, appearance of nonsense-suppressor phenotype for the fusion of the protein and functional domain of Sup35, formation of amyloid aggregates in vitro. (E) Systematic screening for inheritable factors induced by transient overproduction of yeast ORFs. Each approach is illustrated with examples below in the text.