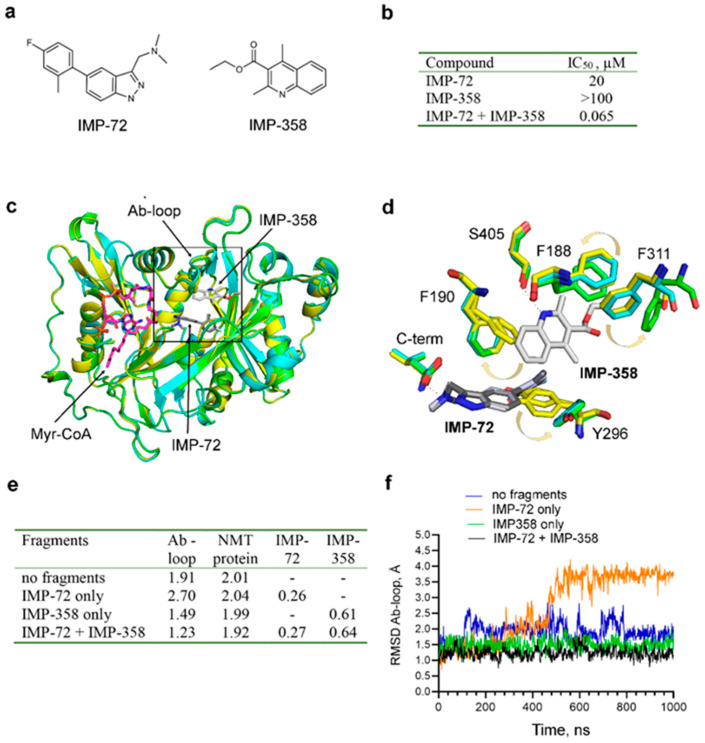
Figure 3.
Fragment synergy could be due to the stabilization of the Ab-loop in its closed state; (a) Structures of IMP-72 and IMP-358, intermediate fragments in the development of IMP-1088; (b) IC50 of IMP-72 or IMP-358 as single agents or IMP-72 in the presence of 100 µM IMP-358. The fragments display remarkable synergism; (c) Structural superimposition of binary PvNMT: Myr-CoA (PDB 4B10, yellow), ternary PvNMT: Myr-CoA: IMP-72 (PDB 5O48, cyan), and quaternary PvNMT: Myr-CoA: IMP-72: IMP-358 (PDB 5O4V, green) complexes. The carbon atoms of IMP-358 are shown in light grey, and those of IMP-72 in a darker shade of grey; (d) A zoomed view of the boxed region in the image on the left. Arrows indicate conformational movements of the depicted residues due to the binding of the fragments. The figure shows that all major conformational changes are proximal and that the binding of IMP-72 does not induce distal conformational changes in the binding pocket of IMP-358 and vice versa. The numbering corresponds to the position of the amino acid residues in HsNMT1. Y296 corresponds to Y211, F188 to F103, F190 to F105, F311 to F226, and S405 to S319 in PvNMT; (e) Average RMSD in angstroms (Å) of the heavy atoms of the Ab-loop, the whole NMT protein, IMP-72, and IMP-358 based on MD simulations of the crystal structure PDB 5O4V, either in the absence of fragments or in the presence of IMP-72, IMP-358, or both of them; (f) RMSD values of the heavy atoms of the Ab-loop in the time course of MD simulations. RMSD < 1.5 Å indicates that the Ab-loop remains locked in its closed conformation by IMP-358.