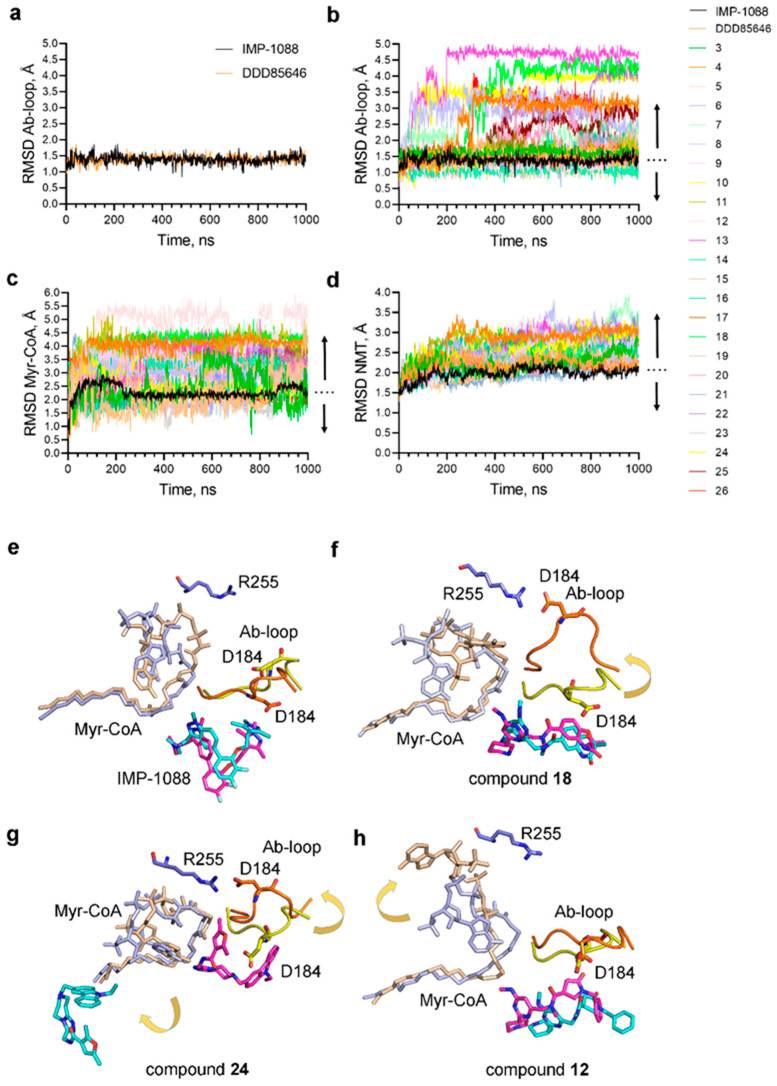
Figure 7.
Conformational changes induced by binding of ligands to HsNMT1 based on MD simulations; (a–d) RMSD of the heavy atoms of the Ab-loop, Myr-CoA, or NMT protein during the time course of MD simulations for the NMT complexes with IMP-1088, DDD85646, and the 24 ligands (compound 3 to compound 26, the legend on the right). RMSD values in the IMP-1088 complexes are indicated in black. The up arrow indicates complexes with increased dynamics (RMSD) compared to the NMT complex of IMP-1088; the down arrow shows complexes with decreased mobility of the indicated Y-axis parameters; (a) The potent NMT inhibitors IMP-1088 and DDD85646 lock the Ab-loop in a closed conformation during the entire duration of MD simulations, RMSD < 1.5 Å; (b) RMSD of the Ab-loop; (c) RMSD of Myr-CoA; (d) RMSD of NMT protein; (e–h) Ab-loop opening and displacement of Myr-CoA during MD simulation of selected NMT-ligand complexes. Structural superimposition of the frames at the beginning (0 ns) and the end (1000 ns) of the MD simulations is shown. In time 0, the ligand is shown in magenta, the Ab-loop in yellow, and Myr-CoA in light blue; in time point 1000 ns, the ligand is shown in cyan, the Ab-loop in orange, Myr-CoA in wheat, and R255 is in blue; (e) In the HsNMT1: IMP-1088 complex, the Ab-loop remains closed, and the cofactor is confined to its binding pocket; (f) In the HsNMT1: compound 18 complex, the Ab-loop opens, leading to the formation of a salt bridge between R255 and D184; (g) In the HsNMT1: compound 24 complex, the Ab-loop opens, and the ligand is completely displaced from the binding site; (h) In the HsNMT1: compound 12 complex, the Ab-loop remains closed, but Myr-CoA is partially displaced from its binding pocket.