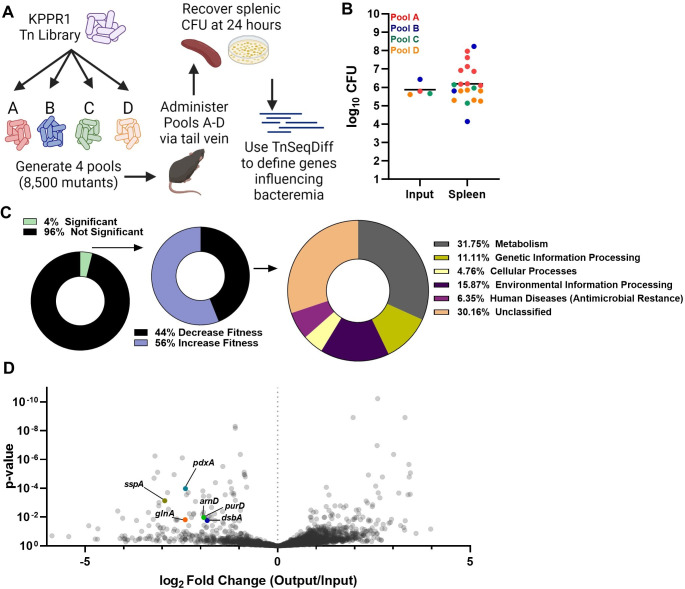
Fig 1. Transposon insertion-site sequencing (TnSeq) reveals that K. pneumoniae bacteremia is enhanced by a set of genes representing diverse fitness mechanisms.
(A) Overview of K. pneumoniae bacteremia TnSeq. A KPPR1 transposon library was divided into four pools containing 8,500 unique insertions and administered to mice via the tail vein at a 1x106 CFU dose. After 24 hours, splenic CFU was recovered, and DNA was sequenced. The TnSeqDiff pipeline determined genes influencing fitness. This illustration was created with BioRender.com. (B) The input and splenic CFU burden for each pool and mouse represented in TnSeq, with mean organ CFU colonization represented by the bar. (C) Genes represented in TnSeq (~3,800 genes) that were defined as influencing infection (132 genes), and primary KEGG orthology for genes increasing K. pneumoniae fitness during bacteremia (58 genes). (D) A volcano plot displaying genes represented in the TnSeq screen with log2 fold change (x-axis) and p-value (y-axis) defined by the TnSeqDiff pipeline. The genes selected for further study, arnD, purD, dsbA, sspA, and pdxA, are highlighted.