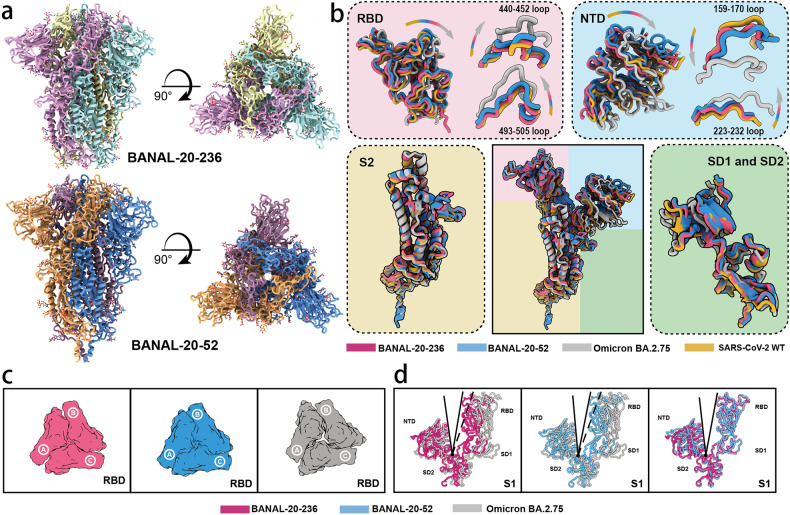
Fig. 2. Cryo-EM structures of the S ectodomain trimers of BANAL-20-52 and BANAL-20-236.
a Cryo-EM map of BANAL-20-52 (PDB ID: 8HXJ) and BANAL-20-236 (PDB ID: 8I3W) S trimer. Three protomers of BANAL-20-236 were colored in yellow, cyan, and magenta, and three protomers of BANAL-20-52 were colored in blue, orange, and pink. Left, side view; right, top view. b The monomeric structures of BANAL-20-236 (magenta) (PDB ID: 8I3W) and BANAL-20-52 (blue) (PDB ID: 8HXJ) S proteins are superimposed to those of SARS-CoV-2 WH strain (PDB ID: 7XU6) (yellow) and Omicron BA.2.75 (PDB ID: 7YQW) (gray). The superimposed individual domain structures and the indicated loops (RBD (top left), NTD (top right), S2 (bottom left) and SD1 and SD2 (bottom right)) are also shown. Both SARS-CoV-2 and omicron BA.2.75 S proteins here are in “down” conformations at an acidic pH. c Top view: Comparison of inter-RBD contacts of the BANAL-20-236 (red), BANAL-20-52 (blue) and Omicron BA.2.75 (gray) S-trimers. d Structural comparison of the angle formed by NTD-SD2-RBD of BANAL-20-236/52 and Omicron BA.2.75. Red, BANAL-20-236; blue, BANAL-20-52; gray, omicron BA.2.75.