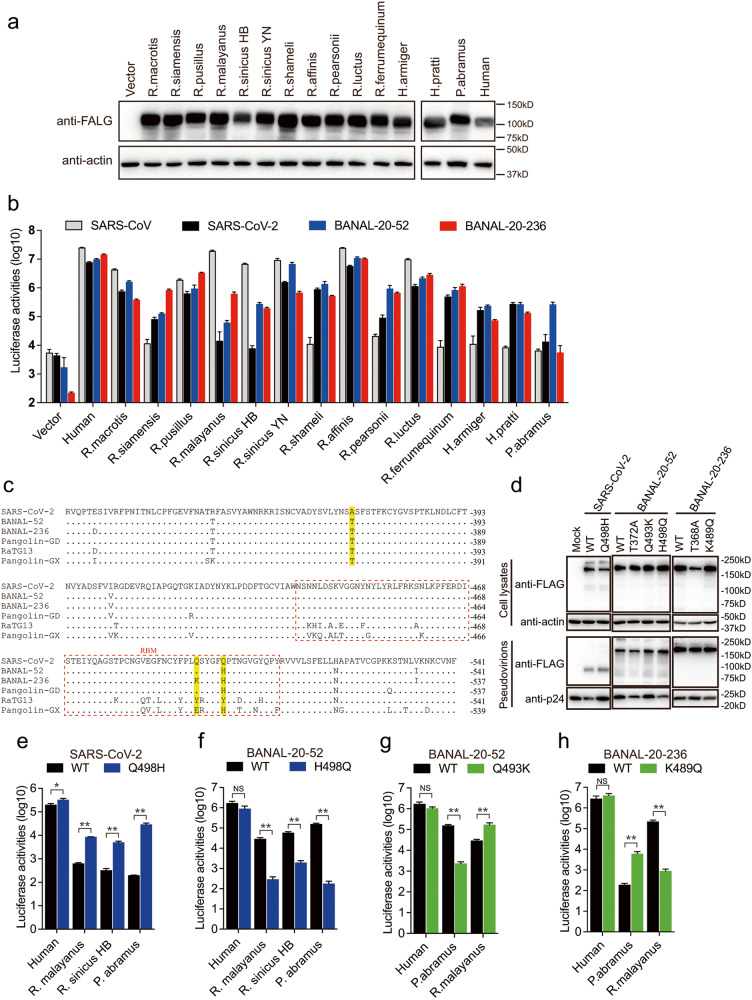
Fig. 3. Susceptibility of different bat ACE2s by BANAL-20-52 S and BANAL-20-236 S pseudovirions.
a Western blotting analysis of cell lysates of HEK293 cells transiently over-expressing FLAG-tagged ACE2 orthologs from different bat species using anti-FLAG M2 antibodies. β-actin served as the loading controls. b Transduction of HEK293 cells expressing ACE2 orthologs from different bat species by SARS-CoV S, SARS-CoV-2 S, BANAL-20-52 S, and BANAL-20-236 S pseudovirions. The experiments were done in triplicate and repeated at least three times. One representative is shown with error bars indicating SD. c Alignment of RBD amino acid sequences from SARS-CoV-2-related sarbecoviruses. RBMs, receptor binding motifs, are indicated by dashed red line box. Residues 372, 493, and 498 are shaded in yellow background. d Detection of SARS-CoV-2 S, BANAL-20-52, or BANAL-20-236 S mutants in cell lysates and pseudovirions by western blot assay using anti-FLAG M2 antibodies. β-actin and gag-p24 served as loading controls (cell lysates, top panel; pseudovirions, bottom panel). e, f Transduction of 293 cells expressing human ACE2, R. malayanus ACE2, R. sinicus HB ACE2, or P. abramus ACE2 by WT and Q498H mutant SARS-CoV-2 S pseudovirions (e), and WT and H498Q mutant BANAL-20-52 S pseudovirions (f). g, h Transduction of 293 cells expressing human ACE2, R. malayanus ACE2, or P. abramus ACE2 by WT and Q493K mutant BANAL-20-52 S pseudovirions (g), and WT and K489Q mutant BANAL-20-236 S pseudovirions (h). Data are represented as means ± SD from at least triplicates. P-values in e–h are calculated by unpaired two-sided Student’s t-test. *P < 0.05; **P < 0.01; ns, P > 0.05.