Figure 1. NOVA1 haploinsufficiency patient and generation of Nova1-deficient mice in Gad2+ inhibitory neurons.
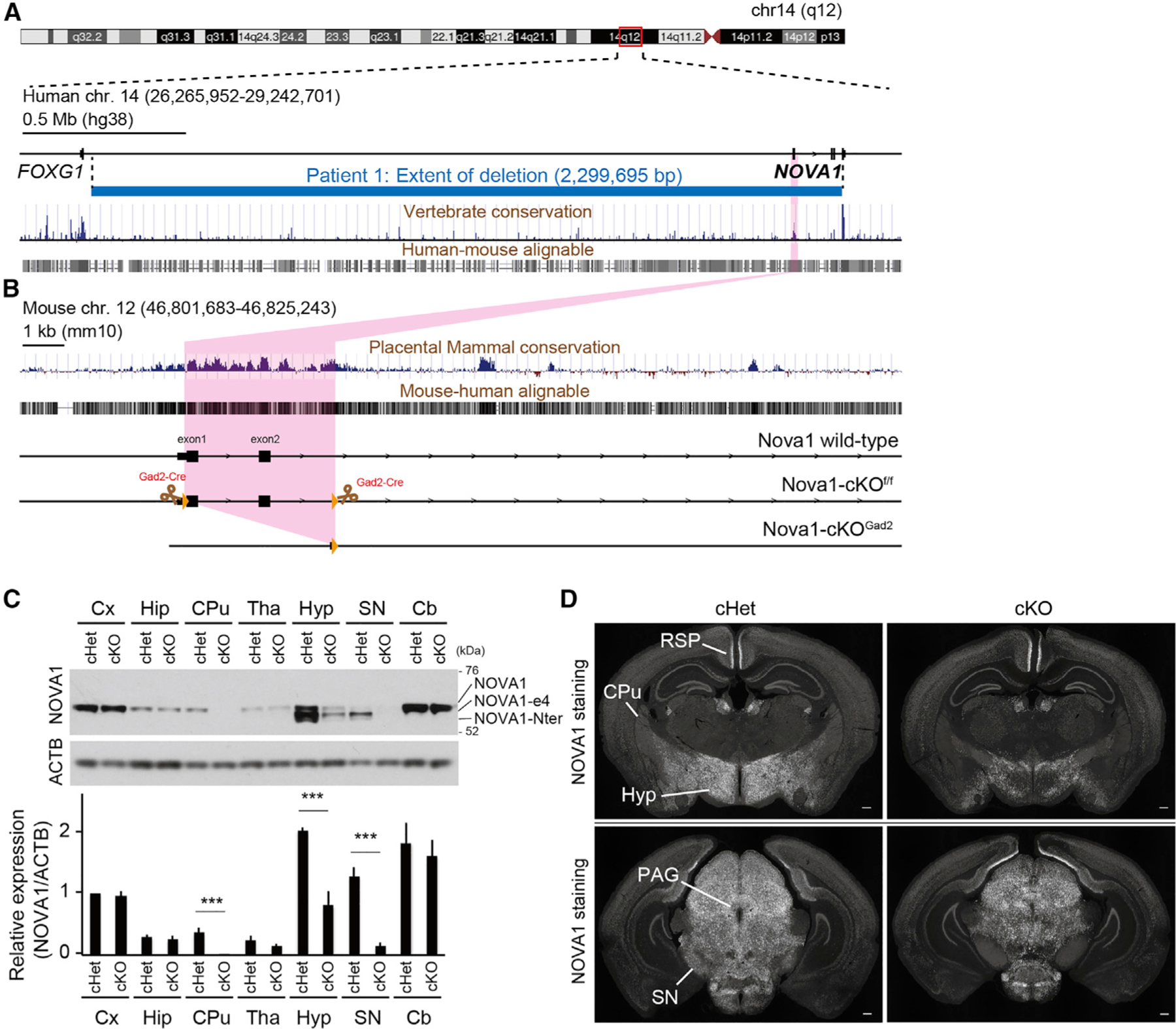
(A and B) Overview of the human (A) and mouse (B) loci. Physical mapping of the deletion at 14q12 (patient) is shown in (A). The detailed view of the region is derived from the University of California, Santa Cruz (UCSC) Genome Browser (GRCh38/hg38 December 2013 assembly). 100-vertebrate PhyloP scores measure vertebrate conservation. Genes are represented in black. Deletion in the patient is depicted by a horizontal blue line. The deletion region for the patient is approximately 2.3 Mb. Nova1-cKOf/f mice were designed with exons 1 and 2 of Nova1 flanked by LoxP sites.
(C) Western blot of NOVA1 in lysates of each dissected brain region of adult mice. NOVA1-e4 is NOVA1 without exon 4, and NOVA1-Nter is an isoform lacking the canonical N terminus. Bottom: quantification of NOVA1 western blot analysis (mean ± SEM, n = 3 biological replicates, unpaired t test, ***p < 0.005).
(D) Immunostaining of P21 cHet (Gad2cre-Nova1f/w) and cKO (Gad2cre-Nova1f/f) mouse brain with NOVA1 antibody. Hyp, hypothalamus; SN, substantia nigra; PAG, periaqueductal gray; CPu, caudate putamen; RSP, retrosplenial region. 40× magnification. Scale bar, 300 μm.
