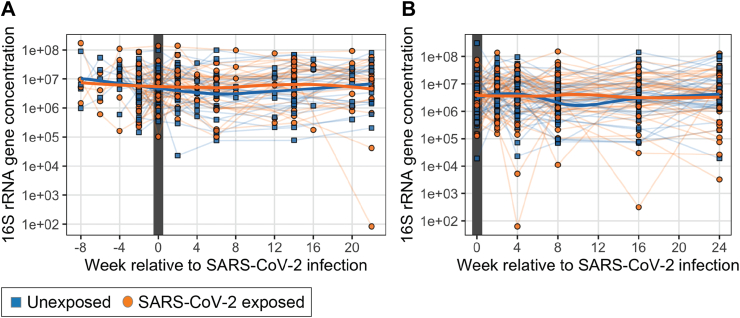
Fig. 2.
Total bacterial populations in the saliva samples in relation to the baseline sample, based on enumeration of 16S rRNA gene copies.A. SARS-CoV-2-infected subjects before, during, and after viral positivity (n = 34) and their 31 matched unexposed counterparts, with the study week shown relative to COVID infection with the week of the first positive test defined as week 0. B. SARs-CoV-2-infected subjects who were virus-positive at the first study visit (n = 47) and their 41 matched unexposed counterparts. There were no significant differences in either analysis between the exposed or unexposed subjects (Student's T test, FDR-corrected p > 0.05).