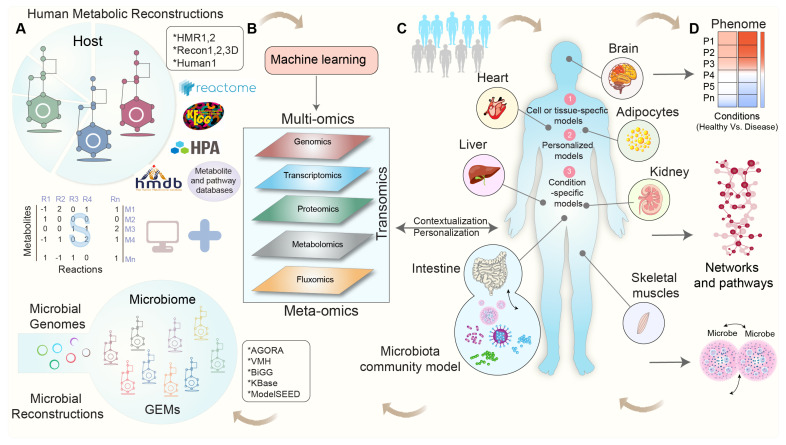
Figure 1.
A schematic workflow for context-specific genome-scale metabolic reconstructions using multi-omics data. (A) Human and microbial metabolic reconstructions and pathway databases serve as a scaffold for integrating omics data, with the stoichiometric matrix (S) representing the mathematical representation of the metabolic network and capturing the stoichiometric coefficients of metabolites (M) in each reaction (R). (B) Various types of multi-omics and/or meta-omics datasets are employed to contextualize human and microbial metabolic reconstructions. (C) Cell-, tissue-, and organ-specific-GEMs are developed using metabolic reconstructions and omics datasets. (D) These condition-specific GEMs enable the predictions of flux phenotypes, the regulation of metabolic pathways, the identification of reporter metabolites and pathways, and the study of microbe-microbe interactions and explore the intricate relationship among diet, host, and microbiota in healthy and disease states.