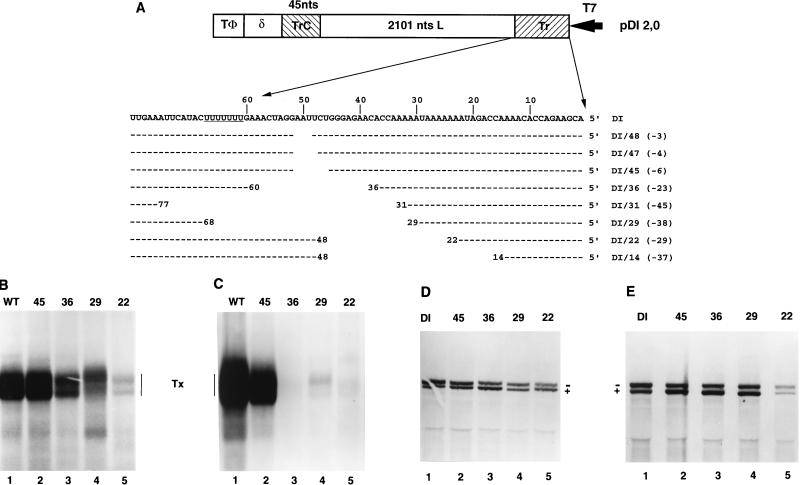
FIG. 6.
Deletions in Tr. (A) Sequences of altered Tr regions of WT and DI-T 5′ deletions. Deletions were generated by using exonuclease III at a unique AflII site located 51 nt in from the genomic 5′ terminus or by site-specific mutagenesis. Each construct is designated WT/X or DI/X, where X is the number of nucleotides at the genomic 5′ terminus that remain. Numbers shown in parentheses are total numbers of nucleotides removed, and positions of nucleotide identity are indicated by dashes. The structure of the WT replicon is shown in Fig. 2; for comparison, the structure of the DI-T replicon is shown here. Note that the deletions were identical for both WT and DI-T. T7, T7 promoter; δ, HDV ribozyme; TΦ, T7 terminator. (B and D) RNA synthesis, determined by agarose-urea gel analysis of VSV-specific RNAs synthesized in cells transfected with the indicated WT (B) or DI-T (D) variants and the VSV N, P, and L support plasmids. RNAs were labeled, harvested, and analyzed as described in the text. Note that the positive (+) and negative (−) strands of DI-T separate under these gel conditions. (C and E) Infectivity assay, determined by agarose-urea gel analysis of VSV-specific RNAs synthesized in cells transfected with the VSV N, P, and L support plasmids and infected with 0.5 ml of the clarified supernatant fluid harvested from transfections that received the VSV N, P, L, M, and G support plasmids and either a WT (C) or DI-T (E) replicon that contained deletions in Tr. Transfections, infections, and RNA analysis were performed as described in Materials and Methods. Tx, products of transcription.