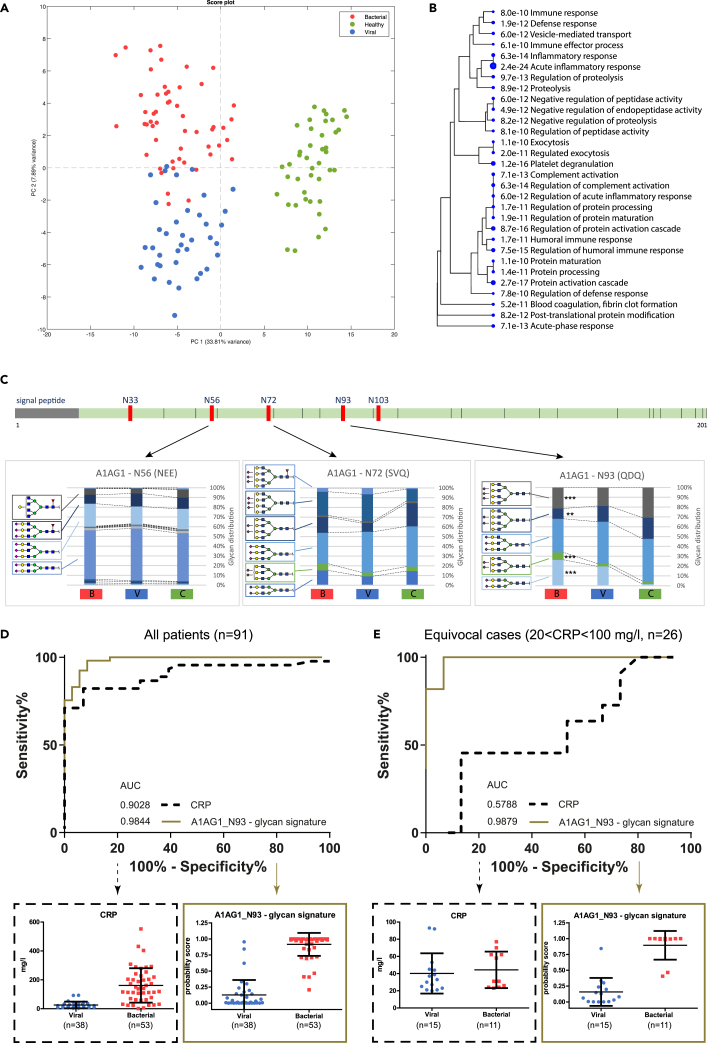
Figure 5.
Separation between bacterial and viral class samples based on fully elucidated glycopeptide features
(A) PCA score plot of 96 identified highly significant features (ANOVA, 99.9% confidence, Bonferroni corrected p < 4.24E-7).
(B) The gene-ontology (GO) hierarchical clustering tree for biological processes summarizing the correlation among the top 30 significant pathways and the corresponding p-value. Pathways with many shared genes/proteins are clustered together. Larger dots indicate higher significant p-values.
(C) Schematic linear representation of α1-acid glycoprotein 1 (AGP or A1AG1) with its five N-glycosylation sites (N33, N56, N72, N93, N103), of which three sites were identified in the dataset. The corresponding cumulative bar graphs show for each N-glycosylation site the measured glycan distribution (%) (y axis), comparing the bacterial [B], viral [V], and healthy control group [C] (x axis), using ANOVA with Bonferroni correction, with ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) ROC curve for all patients (n = 91) based on the clinical C-reactive protein (CRP) levels and the probability score of the model for the glycosylation signature at glycosylation site N93 (A1AG1_N93 – glycan signature). The boxplots below show all individuals for the viral and bacterial group, including mean and standard deviation, based on the CRP levels (left) and the probability score for the glycan signature at site N93 of A1G1 (right).
(E) ROC curve for patients with ambiguous CRP levels (>20 and <100 mg/L, n = 26) showing the clinical CRP and the probability score of the model for the glycosylation signature at glycosylation site N93. The boxplots below show all individuals for the viral and bacterial group, including mean and standard deviation, for the CRP concentration (left) and the probability score for the glycosylation signature at site N93 of A1AG1 (right).