Summary
Here we present a protocol to generate standardized cerebral organoids with hippocampal regional specification using morphogen WNT3a. We describe steps for isolating mouse embryonic (E14.5) neural stem cells from the brain subgranular zone, preparing organoids samples for immunofluorescence, calcium imaging, and metabolic profiling. This protocol can be used to generate mouse brain organoids for developmental studies, modeling disease, and drug screening. Organoids can be obtained in one month, thus providing a rapid tool for high-throughput data validation.
For complete details on the use and execution of this protocol, please refer to Ciarpella et al. “Murine cerebral organoids develop network of functional neurons and hippocampal brain region identity”.1
Subject areas: Cell Biology, Model Organisms, Neuroscience, Stem Cells, Cell Differentiation, Organoids
Graphical abstract

Highlights
-
•
Step-by-step protocol to generate and characterize mouse brain organoids
-
•
Isolation of neural stem cells from the subgranular zone of E14.5 mouse embryos
-
•
Steps to generate mature mouse brain organoids from neural stem cell in 32 days
-
•
Generation of mouse brain organoids with hippocampal signature using WNT3a
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Here we present a protocol to generate standardized cerebral organoids with hippocampal regional specification using morphogen WNT3a. We describe steps for isolating mouse embryonic (E14.5) neural stem cells from the brain subgranular zone, preparing organoids samples for immunofluorescence, calcium imaging, and metabolic profiling. This protocol can be used to generate mouse brain organoids for developmental studies, modeling disease, and drug screening. Organoids can be obtained in one month, thus providing a rapid tool for high-throughput data validation.
Before you begin
Perform all the procedures for the isolation of neural stem cells from mouse embryos under sterile hood as well as all the procedures for cell expansion and the establishment of brain organoids culture. Mouse brain organoids are cultured on an orbital shaker in a humidified incubator at 37°C with 5% CO2. NSCs isolation should take no more than 1 h, and at least 1–2 weeks are necessary for neural stem cells to grow before organoids generation.
The protocol describes the specific steps for using mouse NSCs derived from brain SGZ but the same method can be used to generate brain organoids from meningeal-derived and SVZ-derived NSCs, with comparable results.
Prepare all solutions following the recipes in the materials and equipment section. Some solutions can be prepared in advance and stored as indicated, others must be prepared fresh. Refer to the key resources table for a complete list of materials and equipment.
Institutional permissions
Animal housing and all experimental procedures were approved by the Istituto Superiore di Sanità (I.S.S., National Institute of Health; protocol n. C46F4.N.N4E, Italy) and the Animal Ethics Committee (C.I.R.S.A.L., Centro Interdipartimentale di Servizio alla Ricerca Sperimentale) of the University of Verona (Italy).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SOX2 (1:200) | R&D Systems | Cat# AF2018, RRID: AB_355110 |
| VIMENTIN (1:400) | Millipore | Cat# AB5733, RRID: AB_11212377 |
| KI67 (1:200) | Abcam | Cat# ab16667, RRID: AB_302459 |
| DCX (1:400) | Cell Signaling Technology | Cat# 4604, RRID: AB_561007 |
| B3 TUBULIN (1:400) | Promega | Cat# G7121, RRID: AB_430874 |
| MAP2 (1:200) | Sigma-Aldrich | Cat# M1406, RRID: AB_477171 |
| GFAP (1:200) | Abcam | Cat# ab53554, RRID: AB_880202 |
| SYNAPTOPHYSIN (1:200) | Synaptic Systems | Cat# 101 004, RRID: AB_1210382 |
| ZBTB20 (1:200) | Genetex | Cat# GTX121616, RRID: AB_11177870 |
| KA1 (1:200) | Abcam | Cat# ab67404, RRID: AB_1140942 |
| PSD95 (1:200) | Millipore | Cat# MAB1596, RRID: AB_2092365 |
| GEPHYRIN (1:200) | Synaptic Systems | Cat# 147 011, RRID: AB_887717 |
| VGAT (1:400) | Synaptic Systems | Cat# 131 004, RRID: AB_887873 |
| VGLUT (1:400) | Synaptic Systems | Cat# 135 303, RRID: AB_887875 |
| GAD65/67 (1:400) | Santa Cruz Biotechnology | Cat# sc-365180, RRID: AB_10710523 |
| NMDA (1:400) | Santa Cruz Biotechnology | Cat# sc-365597, RRID: AB_10847218 |
| GLSYN (1:400) | Santa Cruz Biotechnology | Cat# sc-74430, RRID: AB_1127501 |
| GSX2 (1:200) | Genetex | Cat# GTX129390; RRID: AB_288598 |
| FOXG1 (1:200) | Abcam | Cat# ab18259; RRID: AB_732415 |
| NKX2.1 (1:200) | Genetex | Cat# GTX34907 |
| Prealbumin (TTR) (1:200) | Genetex | Cat# GTX85112; RRID: AB_10723946 |
| FRIZZLED 9 (1:200) | Genetex | Cat# GTX71581; RRID: AB_375823 |
| OCT6 (1:200) | Abcam | Cat# ab272925, RRID: AB_2927579 |
| PROX1 (1:200) | Abcam | Cat# ab101851, RRID: AB_10712211 |
| TO-PRO™-3 (1:3000) | Thermo Fisher Scientific | Cat# T3605 |
| Donkey anti-rabbit Alexa Fluor 546 (1:1000) | Thermo Fisher Scientific | Cat# A10040, RRID: AB_2534016 |
| Goat anti-chicken Alexa Fluor 546 (1:1000) | Thermo Fisher Scientific | Cat# A-11040, RRID: AB_2534097 |
| Donkey anti-guinea pig CY3 (1:1000) | Jackson ImmunoResearch | Cat# 706-165-148, RRID: AB_2340460 |
| Goat anti-mouse CY3 (1:1000) | Amersham | Cat# PA43002, RRID: AB_772235 |
| Donkey anti-goat Alexa Fluor 546 (1:1000) | Thermo Fisher Scientific | Cat# A-11056, RRID: AB_142628 |
| Goat anti-rabbit CY3 (1:1000) | Amersham | Cat# PA43004, RRID: AB_772236 |
| Donkey anti-mouse Alexa Fluor 488 (1:1000) | Thermo Fisher Scientific | Cat# A-21202, RRID: AB_141607 |
| Donkey anti-rabbit Alexa Fluor 488 (1:1000) | Thermo Fisher Scientific | Cat# A-21206, RRID: AB_2535792 |
| OXPHOS (1:200) | Abcam | Cat# AB110413, RRID: AB_2629281 |
| VDAC1 (1:200) | Abcam | Cat# 15895, RRID: AB_2214787 |
| Laminin B1 (1:200) | Abcam | Cat# AB16048, RRID: AB_443298 |
| Biological samples | ||
| Embryonic (E14.5) mouse neural stem cells | N/A | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM-F12 GlutaMAX | Gibco | Cat#31331-028 |
| Neurobasal Media | Gibco | Cat#21103-049 |
| Penicillin/Streptomycin (P/S) 100× | Gibco | Cat#15140-122 |
| N-2, 100× | Gibco | Cat#17502-048 |
| B-27, 50× | Gibco | Cat#17504-044 |
| bFGF | Peprotech | Cat#100-18B |
| EGF | Peprotech | Cat#AF-100-15 |
| BDNF | Peprotech | Cat#450-02 |
| WNT3a | Genetex | GTX109037-Pro |
| Accutase | Gibco | Cat#A1105-01 |
| DNAse | Sigma-Aldrich | Cat#D4527 |
| 0.25% Trypsin – EDTA, 1× | Gibco | Cat#25200-056 |
| Trypan blue | Gibco | Cat#15250-061 |
| L-Glutamine 200 mM | Gibco | Cat# 25030081 |
| HBSS 10× | Gibco | Cat#14180-046 |
| HEPES | Sigma | Cat# H3375-250G |
| PFA | Mondial | FM0622 |
| Sucrose | Sigma-Aldrich | Cat#84100 |
| Fetal bovine serum (FBS) | Gibco | Cat# 10270106 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat#472301 |
| Cryobloc (OCT) | Diapath | Cat#070130 |
| Bovine serum albumin (BSA) | Sigma-Aldrich | A7906-10G |
| Triton-X | Sigma-Aldrich | T8787 |
| Sucrose | PanReac AppliChem | A3935 |
| Glucose | Sigma-Aldrich | 47021 |
| Glycerol | Thermo Fisher Scientific | 17904 |
| NaCl | PanReac AppliChem | A2942 |
| KCl | Sigma-Aldrich | P9541 |
| Na2HPO4 | Sigma-Aldrich | S-3234 |
| KH2PO4 | Sigma-Aldrich | P9791 |
| MgCl2-6H2O | J. T. Baker | 0162 |
| CaCl2-2H2O | Sigma-Aldrich | C3881 |
| Tris base | PanReac AppliChem | A2264 |
| EDTA | Sigma-Aldrich | E-5134 |
| Sodium dodecyl sulfate (SDS) | J.T. Baker | 2811 |
| 1,4-Diazabicyclo [2.2.2] octane (DABCO) | Sigma-Aldrich | D-2522 |
| Proteinase K 20 mg/mL | Thermo Fisher Scientific | EO0491 |
| NP-40 | Fluka | 74385 |
| Sodium deoxycholate (DOC) | Sigma-Aldrich | D-6750 |
| Dithiothreitol (DTT) | PanReac AppliChem | A2948 |
| PMSF | Sigma-Aldrich | P7626 |
| Protease inhibitor | Roche | 11836153001 |
| Phosphatase inhibitor | Roche | 04906837001 |
| Critical commercial assays | ||
| Pierce BCA protein assay kit | Thermo Fisher Scientific | 23227 |
| PowerSYBR Green PCR Master Mix | Thermo Fisher Scientific | 4367659 |
| Fluo-4 Direct calcium assay kit | Thermo Fisher Scientific | F10472 |
| Experimental models: organisms/strains | ||
| c57Bl/6J pregnant female mice | Charles River | #001CD |
| Oligonucleotides | ||
| Primer for ND1 forward: 5’ – CTAGCAGAAACAAACCGGGC-3′ | Invitrogen; Quiros et al., 2015 | N/A |
| Primer for ND1 reverse: 5’ – CCGGCTGCGTATTCTACGTT-3′ | Invitrogen; Quiros et al., 2015 | N/A |
| Primer for HK2 forward: 5′-GCCAGCCTCTCCTGATTTTAGTGT-3′ | Invitrogen; Quiros et al., 2015 | N/A |
| Primer for HK2 reverse: 5′- GGGAACACAAAAGACCTCTTCTGG-3′ | Invitrogen; Quiros et al., 2015 | N/A |
| Software and algorithms | ||
| ImageJ | U.S. National Institutes of Health | https://imagej.nih.gov/ij/ |
| Other | ||
| ORBi shaker | Benchmark Scientific | #BT3001 |
| 96-well cell imaging plates | Eppendorf | Cat# 0030741030 |
| 24-well plates | Thermo Fisher Scientific | Cat#144530 |
| Confocal Microscope LSM710 | Zeiss | RRID:SCR_018063 |
| Ti Eclipse Microscope | Nikon | RRID:SCR_021242 |
| NanoDrop™ One/OneC Microvolume UV-Vis Spectrophotometer | Thermo Fisher Scientific | #ND-ONE-W |
| QuantStudio™ 3 Real Time PCR System | Applied Biosystems | N/A |
| Stirrer | Bibby | HB501 |
| pH meter | Crison | Basic20 |
| Centrifuge | Eppendorf | 5417R |
| Centrifuge | MPW | 223e |
| Cryostat | Leica | CM1860 |
| Forceps | FST | 11255–2055 |
| SuperFrost Plus Glass Slides | Epredia™ J1800AMNZ | 10149870 |
| Slides coverslips | Diapath | 061051 |
| Termoblock | FALC | |
| Silicon mold | N/A | N/A |
| Stereomicroscope | N/A | N/A |
| gentleMACS Dissociator | Miltenyi Biotec | |
| gentleMACS C Tubes | Miltenyi Biotec | 130-093-237 |
| Cell strainer 40 mm | Corning | 352340 |
| Syringe filter 0.22 mm | Sartorius | |
For comprehensive key resources table also see Ciarpella et al.1
Materials and equipment
Prepare solution for mouse embryonic (E14.5) SGZ-NSCs isolation
Timing: 1 h
Prepare the solutions necessary for NSCs isolation and culture, as described in the tables below.
Expansion medium without growth factors (can be prepared and stored for 3–4 weeks at 4°C)
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM/F12/Glutamax | N/A | 240 mL |
| P/S, 100× | 1× | 2.5 mL |
| N-2, 100× | 1× | 2.5 mL |
| B-27, 50× | 1× | 5 mL |
| Total | 250 mL |
CRITICAL: Sterilize by filtering with a 0.22 μm filter.
Note: The suitability of the solution can be quickly check by looking at the color of the media. DMEM/F12/Glutamax contains Phenol Red thus providing indication about pH solution. Under suitable condition, growth medium will have a warm pink-red color. After long storage, the indicator color turns to bright pink. If contamination occurred during storage, the indicator color may turn into yellow.
Expansion Medium (to be prepared fresh)
| Reagent | Final concentration | Amount |
|---|---|---|
| Expansion medium | N/A | 50 mL |
| bFGF | 20 ng/mL | 50 μL |
| EGF | 20 ng/mL | 50 μL |
| Total | 50 mL |
HEPES (can be prepared and stored for 3–4 weeks at 4°C) pH 7.3
| Reagent | Final concentration | Amount |
|---|---|---|
| HEPES | 0.3 M | 7.15 g |
| dH2O | N/A | 80 mL |
| Total | 100 mL |
CRITICAL: check and adjust the pH before each use; sterilize by filtering with a 0.22 μm filter.
HBSS 1× (can be prepared and stored for 3–4 weeks at 4°C) pH 7.4
| Reagent | Final concentration | Amount |
|---|---|---|
| dH20 | N/A | 428.5 mL |
| HBSS 10× | 1× | 50 mL |
| 0.3 M HEPES, pH 7.3 | 0.01 M | 16.5 mL |
| P/S, 100× | 1× | 5 mL |
| Total | 500 mL |
CRITICAL: check and adjust the pH before use; sterilize by filtering with a 0.22 μm filter.
Note: The suitability of the solution can be quickly check by looking at the color of the media. HBSS 10× contain Phenol Red thus providing indication about pH solution. Under suitable condition, growth medium will have a warm pink-red color. After long storage, the indicator color turns to bright pink. If contamination occurred during storage, the indicator color may turn into yellow.
PBS 10× (can be prepared and stored for 3–4 weeks at 4°C)
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl | 1.37 M | 80 g |
| KCl | 27 mM | 2 g |
| Na2HPO4 | 81 mM | 11.5 g |
| KH2PO4 | 14.7 mM | 2 g |
| dH2O | N/A | ∼1000 mL |
| Total | 1000 mL |
CRITICAL: Sterilize by filtering with a 0.22 μm filter.
Note: PBS 10× is a stock solution. PBS 1× should be prepared by dilution in dH2O.
SGZ-NSCs Extraction Solution (to be prepared the day before use and stored at 4°C, trash out any solutions’ residual after usage)
| Reagent | Final concentration | Amount |
|---|---|---|
| Trypsin/EDTA 1× | 0.2× | 600 μL |
| DNAse, 120 U/mL | 0.4 U/mL | 10 μL |
| PBS 1× | N/A | 2390 μL |
| Total | 3 mL |
Note: The final total volume of 3 mL of “SGZ-NSCs Extraction solution” is necessary and sufficient for one litter. Adjust the volume of solution accordingly to litters that have to be used.
Prepare solution for mouse brain organoids generation and further analysis
Timing: 1 h
All the solutions necessary for mouse brain organoids generation, culture maintenance and further processing are described in the tables below.
Expansion medium without growth factors (can be prepared and stored for 3–4 weeks at 4°C)
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM/F12/Glutamax | N/A | 240 mL |
| P/S, 100× | 1× | 2.5 mL |
| N-2, 100× | 1× | 2.5 mL |
| B-27, 50× | 1× | 5 mL |
| Total | 250 mL |
CRITICAL: Sterilize by filtering with a 0.22 μm filter.
Note: The suitability of the solution can be quickly check by looking at the color of the media. DMEM/F12/Glutamax contains Phenol Red thus providing indication about pH solution. Under suitable condition, growth medium will have a warm pink-red color. After long storage, the indicator color turns to bright pink. If contamination occurred during storage, the indicator color may turn into yellow.
Neuron Chow without differentiation factor (can be prepared and stored for 3–4 weeks at 4°C)
| Reagent | Final concentration | Amount |
|---|---|---|
| Neural Basal Medium | N/A | 242 mL |
| P/S, 100× | 1× | 2.5 mL |
| L-Glutamine 200 mM | 4 mM | 500 μL |
| B-27, 50× | 1× | 5 mL |
| Total | 250 mL |
CRITICAL: Sterilize by filtering with a 0.22 μm filter.
Note: The suitability of the solution can be quickly check by looking at the color of the media. Neural Basal Medium contains Phenol Red thus providing indication about pH solution. Under suitable condition, growth medium will have a warm pink-red color. After long storage, the indicator color turns to bright pink. If contamination occurred during storage, the indicator color may turn into yellow.
Expansion Medium (to be prepared fresh) (for brain organoids culture from day 0 to day 4)
| Reagent | Final concentration | Amount |
|---|---|---|
| Expansion medium | N/A | 50 mL |
| bFGF | 20 ng/mL | N/A |
| EGF | 20 ng/mL | N/A |
| (optional WNT3a) | (5 ng/mL) | (N/A) |
| Total | 50 mL |
Induction medium I (to be prepared fresh) (for brain organoids culture from day 5 to day 6)
| Reagent | Final concentration | Amount |
|---|---|---|
| Expansion medium | N/A | 50 mL |
| bFGF | 10 ng/mL | N/A |
| EGF | 10 ng/mL | N/A |
| (optional WNT3a) | (5 ng/mL) | (N/A) |
| Total | 50 mL |
Induction medium II (to be prepared fresh) (for brain organoids culture from day 7 to day 14)
| Reagent | Final concentration | Amount |
|---|---|---|
| Expansion medium | N/A | 50 mL |
| bFGF | 5 ng/mL | N/A |
| (optional WNT3a) | (5 ng/mL) | (N/A) |
| Total | 50 mL |
Differentiation media (to be prepared fresh) (for brain organoids culture from day 15 to day 32)
| Reagent | Final concentration | Amount |
|---|---|---|
| Neuron Chow | N/A | 50 mL |
| BDNF | 50 ng/mL | N/A |
| Total | 50 mL |
Fixing solution (to be prepared fresh)
| Reagent | Final concentration | Amount |
|---|---|---|
| PFA | 4% | 100 mL |
| Sucrose | 4% | 4 g |
| Total | 100 mL |
CRITICAL: PFA is hazard and the “Fixing solution” must be prepared under a chemical hood.
Triton-X stock solution (0.5%) (can be prepared and stored for 2–3 months at 4°C)
| Reagent | Final concentration | Amount |
|---|---|---|
| Triton-X | 0.5% | 5 mL |
| PBS 1× | N/A | 995 mL |
| Total | 1000 mL |
CRITICAL: Triton-X is viscous, a p1000 cut tip should be used to draw it. Triton-X takes time to be solubilized, use a magnetic stirrer and leave the tip inside the bottle to allow complete Triton-X dissolution.
Blocking solution 0.5% (to be prepared fresh) (for nuclear marker staining)
| Reagent | Final concentration | Amount |
|---|---|---|
| BSA | 2% | 0.2 g |
| Triton-X stock solution | 0.5% | 10 mL |
| Total | 10 mL |
CRITICAL: BSA takes minutes to be solubilized, use a magnetic agitator to ensure proper dissolution. Keep on ice during usage.
Blocking solution 0.25% (to be prepared fresh) (for cytosolic marker staining)
| Reagent | Final concentration | Amount |
|---|---|---|
| BSA | 2% | 0.2 g |
| Triton-X stock solution | 0.25% | 5 mL |
| PBS 1× | N/A | 5 mL |
| Total | 10 mL |
CRITICAL: BSA takes minutes to be solubilized, use a magnetic agitator to ensure proper dissolution. Keep on ice during usage.
DABCO stock (prepare and store 2–3 months at 4°C)
| Reagent | Final concentration | Amount |
|---|---|---|
| DABCO powder | 10 mg/mL | 10 mg |
| dH2O | N/A | 1 mL |
| Total | 1 mL |
CRITICAL: DABCO is light sensitive, wrap the Eppendorf in tin foil and keep away from light.
DABCO solution (to be prepared fresh and stored 3–4 weeks at 4°C)
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS 1× | N/A | 5 mL |
| Glycerol 100% | 50% | 5 mL |
| DABCO stock | 0.1 mg/mL | 100 μL |
| Total | 10 mL |
CRITICAL: DABCO is light sensitive, wrap the falcon in tin foil and keep away from light. Do not use it if the solution remains at 20°C–22°C for more than 2 h.
PSS solution (can be prepared and stored 3–4 weeks at 4°C)
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl | 140 mM | 4 g |
| KCl | 5 mM | 0.18 g |
| Na2HPO4 | 1.2 mM | 0.08 g |
| MgCl2-6H2O | 1.4 mM | 0.14 g |
| CaCl2-2H2O | 1.8 mM | 0.13 g |
| Glucose | 11.5 mM | 1.03 g |
| HEPES | 10 mM | 1.19 g |
| Total | 500 mL |
CRITICAL: Check and adjust the pH before use; sterilize by filtering with a 0.22 μm filter.
Lysis buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| TrisHCl pH 8.5–8.8 1 M | 100 mM | 100 mL |
| EDTA 0.5 M | 5 mM | 10 mL |
| SDS 10% | 0.2% | 20 mL |
| NaCl 5 M | 200 mM | 40 mL |
| dH2O | N/A | 830 mL |
| Total | 1000 mL |
Proteinase K solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Proteinase K 20 mg/mL | 10 mg/mL | 500 μL |
| Glycerol 100% | 25% | 250 μL |
| dH2O | N/A | 250 μL |
| Total | 1 mL |
PCR working solution (volume for 1 sample in single replicate)
| Reagent | Final concentration | Amount |
|---|---|---|
| Master Mix 2× | 1× | 10 μL |
| Primers F and R 100 μM | 10 μM | 0,1 μL |
| DNAse Free Water | N/A | 5,8 μL |
| Total | 16 μL |
RIPA Buffer (can be prepared and stored 2–3 weeks at 4°C; 2–3 months at −20°C)
| Reagent | Final concentration | Amount |
|---|---|---|
| TrisHCl 1 M pH 7.4 | 50 mM | 500 μL |
| NaCl 5 M | 100 mM | 200 μL |
| NP-40 | 1% | 50 μL |
| Sodium deoxycholate (DOC) 10% | 1% | 1 mL |
| MgCl2 1 M | 5 mM | 50 μL |
| Glycerol 100% | 3% | 300 μL |
| dH2O | N/A | 7.9 mL |
| Total | 10 mL |
Note: The detergents in the RIPA buffer may precipitate over time. If this happens, heat the solution (37°C) and mix to dissolve the components.
Complete RIPA BUFFER (to be prepared fresh and used right away)
| Reagent | Final concentration | Amount |
|---|---|---|
| Dithiothreitol (DTT) 0.5 M | 5 mM | 10 μL |
| PMSF (stock 200 mM in EtOH 100%) | 5 mM | 25 μL |
| Protease inhibitor 7× | 1× | 142.9 μL |
| Phosphatase inhibitor 10× | 1× | 100 μL |
| RIPA buffer | 1× | 722.1 μL |
| Total | 1 mL |
Step-by-step method details
Mouse embryonic (E14.5) neural stem cells isolation from SGZ
Timing: 1 h
The starting point for the generation of mouse brain organoids is the isolation of neural stem cells (NSCs) from the subgranular zone (SGZ), a well-known stem cells niche.2,3 The peak of neurogenesis of mouse brain is at embryonic day 14.5 (E14.5)4,5 therefore to ensure an efficient isolation of NSCs we extract cells at this developmental stage.
CRITICAL: tissue harvesting, and cells isolation procedures should take no more than 1 h to ensure a high yield of viable stem cells.
Note: This protocol is based on the use of c57Bl/6J mouse colonies. Similar results could be obtained with other mouse colonies (e.g. CD1 mice).
-
1.
Sacrifice pregnant adult female mouse by cervical dislocation.
Note: to ensure the collection of embryos at E14.5 it is important to determine the day 0 of pregnancy by following specific procedure6 or by relying on vendor information.
CRITICAL: cervical dislocation must be performed by trained and qualified personnel only. Cervical dislocation without prior anesthesia with CO2 or isofluorane inhalation ensures high yield of healthy, viable embryos7,8 however a fast CO2 narcosis or isoflurane induction before the cervical dislocation can be performed.
-
2.Clean the mouse skin with Et-OH 70% and quickly harvest the embryos.
-
a.Grab skin below the center of the belly with forceps and cut through the skin to expose the abdominal and thoracic cavities.
-
b.Remove the sac containing the string of embryos.
-
c.Remove the embryos one-by-one by gently cutting away the outer membrane (Figure 1A).
-
a.
-
3.
Collect the embryos’ heads in HBSS 1× on ice.
Note: hereafter work under sterile hood.
-
4.
Under stereomicroscope isolate the brain, divide the hemispheres and process one-by-one (Figures 1B–1D, Methods video S1).
CRITICAL: keep the half hemispheres hydrated with HBSS 1× during the procedure to avoid tissue degradation.
-
5.
Identify the SGZ region (Figures 1E and 1F), carefully isolate the zone by using forceps.
Note: at this embryonic stage, the hippocampal region is not completely formed and thus the dentate gyrus and the SGZ is not clearly visible.
Figure 1.
Representative images of embryonic (E14.5) mouse brain to facilitate SGZ recognition and dissection
(A) Procedure of embryos extraction.
(B) Coronal view of intact brain extracted from E14.5 mouse embryo. Both hemispheres and a portion of brainstem are visible. Cut off the brainstem to proceed only with the hemispheres.
(C) The hemispheres are separated by cutting at the middle. Red dashed lines in B-C indicate the cutting lines.
(D) One by one hemisphere is flipped 90° side down. The flipped hemisphere presents a tissue hump (delimitated by green dashed line and black head arrow) representing the midbrain and thalamus inner brain regions.
(E) Scheme of a sagittal section of E14.5 mouse brain highlighting the anatomical structures. The hippocampal allocortex (delimitated by blue dashed line) hosts the primordium of SGZ.
(F) The SGZ resides behind the midbrain and thalamus inner brain tissue; carefully lift it to harvest the SGZ. LH: left hemisphere, RH: right hemisphere; SGZ: sub granular zone.
Refer to Methods video S1 for the procedure of SGZ isolation.
-
6.
Collect the extracted tissues in a 15 mL falcon tube, previously filled with 6 mL of HBSS and keep it on ice (4°C).
-
7.
Repeat step 4–6 for each embryos’ brain and pool all the harvested SGZs together.
-
8.
Centrifuge 1 min 300 g and discard the supernatant solution by using pipette.
CRITICAL: the tissue pellet is faint, do not turn the falcon tube to remove the supernatant.
-
9.
Wash the tissue pellet with PBS 1×.
-
10.
Repeat step 8.
-
11.
Resuspend the tissue pellet in 3 mL of SGZ-NSCs extraction solution by gently mixing.
-
12.
Transfer the suspension in a gentleMACS tube (Purple-cap C Tubes – for tissue dissociation) (Miltenyi Biotec).
-
13.
Insert the tube into the gentleMACS Dissociator instrument, running the following pre-set gentleMACS Program: BRAIN_01_01′.
-
14.
Incubate the tube at 37°C water bath for 10 min.
-
15.Insert the tube into the gentleMACS Dissociator instrument.
-
a.Running the pre-set gentleMACS Program BRAIN_02_01′.
-
b.Running the pre-set gentleMACS Program BRAIN_03_01′.
-
a.
CRITICAL: if the tissue looks not completely homogenate, repeat once again step 15a-b.
Alternatives: if the gentleMACS Dissociator instrument is not available, tissue dissociation can be performed by manual mechanical dissociation: vigorously pipette up and down (at least 20 times) the solution by using a p1000 and then 10 times by using a Pasteur pipette. Note that this procedure could result in a high number of dead cells.
-
16.
Filter the solution with a 40 μm cell strainer in a 50 mL falcon tube.
-
17.
Centrifuge 10 min 300 g, gently remove the supernatant by using a pipette.
-
18.
Resuspend the pellet in 1 mL of Expansion medium to obtain the cell suspension.
-
19.Count cells:
-
a.Mix 90 μL of Trypan Blue with 10 μL of the cell suspension (step 18) in a 500 μL Eppendorf.
-
b.Put 10 μL of the suspension prepared in a. into the Burker counting chamber.
-
c.Count viable cells by using a bright-field inverted microscope.
-
a.
Note: cell count can be performed with the preferred method; automatic cell counter or different chambers e.g., Neubauer chamber can be used.
-
20.
Seed 500′000 viable cells in a 75 cm2 flask filled with 12 mL of Expansion medium.
CRITICAL: low yield of cells could be obtained from isolation due to few starting embryos or to an incomplete tissue dissociation. Seed the cells in 25 cm2 flask or 6-wells plate accordingly to the number of cells obtained.
-
21.
Culture the NSCs in incubator 37°C until the formation of neurospheres.
CRITICAL: daily examinate the NSCs culture under microscope to check potential contamination.
-
22.
Refill the cells medium with 1 mL of fresh Expansion medium every other day.
Note: at least 1–2 weeks are necessary for neural stem cells neurospheres to reach the right density before splitting them (see steps 23–34).
Neurospheres splitting and expansion
Timing: 20 min
This section describes the procedure to enzymatically dissociate neurospheres to obtain single cells necessary to generate organoids. Single cells can be expanded and maintained in culture or even frozen.
-
23.
Collect the medium containing neurospheres into a 15 mL falcon tubes.
CRITICAL: check under the microscope if any neurospheres are still present in the flask to ensure to collect most of the cells.
Note: if a lot of neurospheres are still visible in the flask, wash it with PBS 1× ensuring to harvest almost all of them.
-
24.
Centrifuge 5 min at 300 g.
-
25.Carefully discard the supernatant by using a 10 mL serological pipette.
-
a.Use p1000/p200 tip to completely remove any liquid residual.
-
a.
CRITICAL: excess of liquid will affect the Accutase dissociation.
-
26.
Add 200 μL of Accutase to the pellet.
Note: 200 μL of Accutase is used for a cell pellet derived from 2 T75 flasks.
-
27.
Gently homogenize the pellet with Accutase for max 3–5 min avoiding bubbles formation.
Note: stop the Accutase process as soon as the solution become opaque and neurosphere aggregates are no longer visible to unaided eye (see Figure 2).
CRITICAL: long Accutase treatment or aggressive pipetting may cause cell death.
-
28.
Add PBS 1× up to 6 mL to dilute the Accutase.
-
29.Centrifuge 5 min at 300 g.
-
a.Carefully discard the supernatant by using a 10 mL serological pipette.
-
b.Use p1000/p200 tip to completely remove any liquid residual.
-
a.
-
30.
Resuspend the single cells pellet in 1 mL of Expansion medium to obtain cell suspension.
-
31.Cell count:
-
a.Mix 90 μL of Trypan Blue with 10 μL of the cell suspension (step 30) in a 500 μL Eppendorf.
-
b.Put 10 μL of the suspension prepared in a. into the Burker counting chamber.
-
c.Count viable cells by using a bright-field inverted microscope.
-
a.
Note: 3 millions of cells are generally obtained from a single T75 flask. If high number of cells is obtained, NSCs can be frozen (step 33) or used to further expansion (step 34).
-
32.
At this step cells can be directly used in step 35 to generate organoids following unguided protocol or in step 38 to generate organoids with for hippocampal specification.
-
33.(Optional) Resuspend the cells to be frozen in at least 1 mL of 50% expansion medium and 50% of freezing solution (20% DMSO plus 80% FBS).
-
a.Quick store the vial at −20°C for few hours to let the cells freeze.
-
b.Move the frozen cells at −80°C.Note: frozen cells can be stored at −80°C for several months.
-
c.Move the frozen cells in liquid azote for long (years) storage.
-
a.
-
34.(Optional) Plate 500′000 viable cells or 250′000 viable cells in T75 or T25 flask, respectively.
-
a.Supplement the T75 or T25 flask with 12 mL or 6 mL of Expansion medium, respectively.
-
b.Refill with 1 mL or 500 μL fresh Expansion medium every other day, until neurospheres forming.
 CRITICAL: avoid repeated splitting and expansion of cells as neurospheres to prevent NSCs differentiation and attachment to the plate. The maximum number of splitting recommended is 4–5.
CRITICAL: avoid repeated splitting and expansion of cells as neurospheres to prevent NSCs differentiation and attachment to the plate. The maximum number of splitting recommended is 4–5.
-
a.
Figure 2.
Representative images of neurospheres to be processed for Accutase treatment
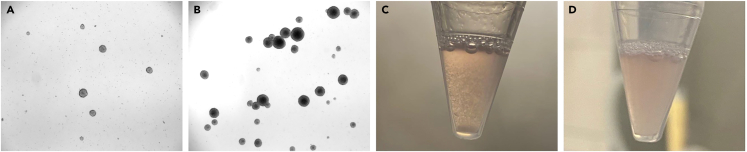
The majority of cells inside the Flask should be as neurospheres before splitting. Once the Accutase is added to cell pellet, gently pipetting up and down the cellular suspension until the solution became milky and neurospheres are no longer visible.
(A) Neurospheres not ready to be splitted.
(B) Neurospheres ready to be splitted.
(C) Cell suspension before Accutase treatment.
(D) Cell suspension after Accutase treatment.
Brain organoid culture (unguided protocol)
Timing: 1 month
This section describes the three phases (see Figure 3) that lead to the generation of mouse brain organoids with consistent size, organized neural network and functional neurons within 32 days. The organoid generation method described in this section relies on spontaneous neuronal differentiation without any extrinsic patterning to guide the development into a specific brain region. However, due to the neural stem cell source used, the organoids may have an intrinsic patterning toward dorsal, rather than ventral, brain region specification. To generate organoids with hippocampal-specific phenotype directly go to step 38.
Note: Keep brain organoids in dynamic culture throughout all the phases (ORBi shaker at 75rpm).
-
35.Expansion phase (days 0‒4):
-
a.Day 0: seed 20′000 single viable cells/well (day 0) on 24 well plates in 500 μL of culture medium Expansion medium.
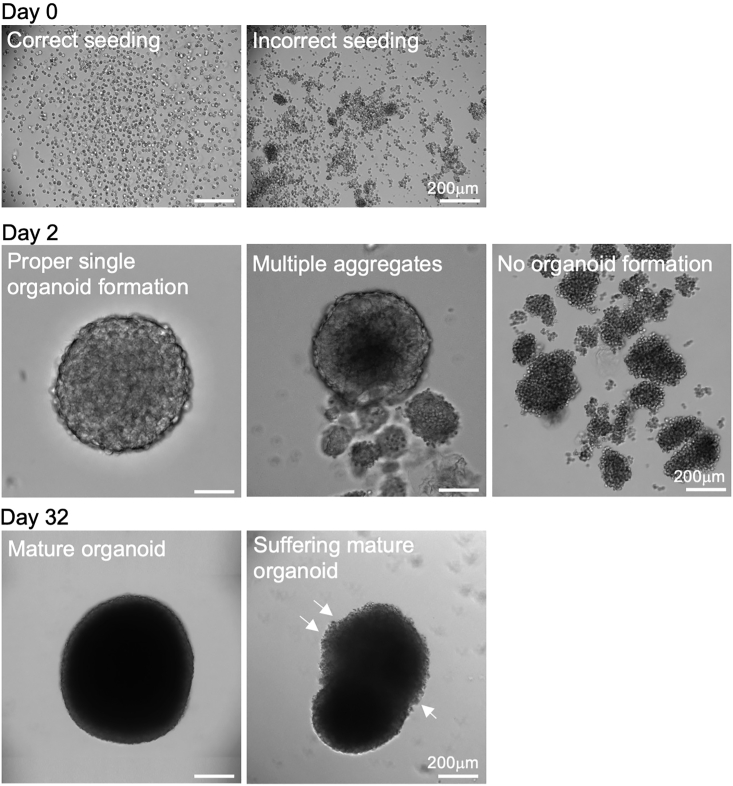
 CRITICAL: seeding must be homogeneous for the organoids to form in a proper way (see Figure 4). Ensure pipetting before each seed. Troubleshooting 1
Note: seed the cells only in 18 wells of the 24 well plate can help to maintain the humidity of the plate and better allow the growth of the brain organoids. Putting sterile PBS 1× into the last row of the plate.
CRITICAL: seeding must be homogeneous for the organoids to form in a proper way (see Figure 4). Ensure pipetting before each seed. Troubleshooting 1
Note: seed the cells only in 18 wells of the 24 well plate can help to maintain the humidity of the plate and better allow the growth of the brain organoids. Putting sterile PBS 1× into the last row of the plate. -
b.Day 1: check cells assembly under a microscope. Troubleshooting 2.
-
c.Day 2: discard 80 μL of the culture medium and refill with 100 μL of fresh Expansion medium.
-
d.Day 4: discard 80 μL of the culture medium and refill with 100 μL of fresh Expansion medium.
-
a.
-
36.Induction phase (days 5‒14): to promote spontaneous NSCs differentiation, gradually lower growth factors’ concentration.
-
a.Day 5: completely replace Expansion medium with 500 μL of Induction medium I.
 CRITICAL: at this stage, brain organoids are fragile, and it is recommended to avoid touching them during the medium change. To ease the medium change, put a black sheet under the plate to better visualize the samples and tilt the plate to let the brain organoids lay on one side of the well.
CRITICAL: at this stage, brain organoids are fragile, and it is recommended to avoid touching them during the medium change. To ease the medium change, put a black sheet under the plate to better visualize the samples and tilt the plate to let the brain organoids lay on one side of the well. -
b.Day 7: completely replace Induction medium I with 500 μL of Induction medium II.
 CRITICAL: To ease the medium change, put a black sheet under the plate to better visualize the samples and tilt the plate to let the brain organoids lay on a side.
CRITICAL: To ease the medium change, put a black sheet under the plate to better visualize the samples and tilt the plate to let the brain organoids lay on a side. -
c.Day 9: discard 80 μL of the medium and refill with 100 μL of fresh Induction medium II every other day (on day 11 and day 13).
-
a.
-
37.Differentiation phase (days 15‒32): boost brain organoids’ neural differentiation by supplementing the culture medium with BDNF.
-
a.Day 15: completely replace Induction medium II with 500 μL of Differentiation medium.
 CRITICAL: at this stage, brain organoids are small and they could be accidentally taken up. To ease the medium change, put a black sheet under the plate to better visualize the samples and tilt the plate to let the brain organoids lay on a side.
CRITICAL: at this stage, brain organoids are small and they could be accidentally taken up. To ease the medium change, put a black sheet under the plate to better visualize the samples and tilt the plate to let the brain organoids lay on a side. -
b.Day 17: discard 80 μL of the medium and refill with 100 μL of fresh Differentiation medium every other day (on days 19, 21, 23, 25, 27, 29, 31).Note: Brain organoids characterization can be performed at different time points (i.e., day 7, day 14, day 21 and day 32) to follow the cellular organization and the differentiation progress. To do that, some of the organoids kept in culture can be appropriately stored at the desired time for further analysis (see step 41 for immunofluorescence analysis; step 79 for calcium imaging; step 85 for mtDNA analysis; step 107 for protein extraction).Representative images of brain organoids morphology at different stages of the culture protocol are reported in Figure 5.
-
a.
Figure 3.
Culturing steps of mouse brain organoids protocol and related media and factors used
The protocol consists of three steps: expansion (days 0 to 4), induction (days 5 to 14) and differentiation (days 15 to 32) phase. In each phase, the organoids are provided with media enriched with defined growth (bFGF and/or EGF) or differentiation (BDNF) factors. For hippocampal phenotype induction the morphogen WNT3a is added to culture media during the expansion and induction phase.
Figure 4.
Representative organoids images at critical step of the protocol. Scale bars: 200μm
Figure 5.
Organoids morphology at different stages of the culture protocol. Scale bars: 200μm
Hippocampal phenotype specification in brain organoids
Timing: 1 month
This section describes the generation of mouse brain organoids with hippocampal-specific phenotype.
The hippocampus is one of the brain regions mostly involved in the pathology progression of many neurodegenerative disorders including Alzheimer’s disease and epilepsy.9,10 By adding the WNT3a factor, crucial for the hippocampal fate determination,11 it is possible to generate mouse brain organoids that specifically own hippocampal signature. To this end, the three steps of the brain organoid culture are maintained (steps 35–37) (see Figure 3), with slight modification on the media provided during the expansion and induction phase. Specifically, 5 ng/mL WNT3a is added to Expansion medium, Induction medium I and Induction medium II. Key passages are reported below.
Note: Keep brain organoids in dynamic culture throughout all the phases (ORBi shaker at 75 rpm).
-
38.Expansion phase (days 0‒4):
-
a.Day 0: seed 20′000 single viable cells/well (day 0) on 24 well plates in 500 μL of Expansion medium plus 5 ng/mL WNT3a. Troubleshooting 1.
-
b.Day 1: check cells assembly under microscope. Troubleshooting 2.
-
c.Day 2: discard 80 μL of the culture medium and refill with 100 μL of fresh Expansion medium plus 5 ng/mL WNT3a.
-
d.Day 4: discard 80 μL of the culture medium and refill with 100 μL of fresh Expansion medium plus 5 ng/mL WNT3a.
-
a.
-
39.Induction phase (days 5‒14): to promote spontaneous NSCs differentiation, gradually lower growth factors’ concentration.
-
a.Day 5: completely replace Expansion medium with 500 μL of Induction medium I plus 5 ng/mL WNT3a.
-
b.Day 7: completely replace Induction medium I with 500 μL of Induction medium II plus 5 ng/mL WNT3a.
-
c.Day 9: discard 80 μL of the medium and refill with 100 μL of fresh Induction medium II plus 5 ng/mL WNT3a every other day (on day 11 and day 13).
-
a.
-
40.Differentiation phase (days 15‒32): boost brain organoids neural differentiation by supplementing the culture medium with BDNF.Note: hereafter, addition of Wnt3a is not necessary anymore.
-
a.Day 15: completely replace Induction medium II with 500 μL of Differentiation medium.
-
b.Day 17: discard 80 μL of the medium and refill with 100 μL of fresh Differentiation medium every other day (on day 19, 21, 23, 25, 27, 29, 31).We did not report any statistically significant differences in the morphology of brain organoids with hippocampal signature and the ones generated with the unguided protocol when observed in brightfield microscopy (see Figure 6). Likewise, we didn’t find any significant differences in size between the two set of brain organoids (see Figure 6). However, by adding WNT3a at low concentration (5 ng/mL) you will be able to obtain hippocampal specific neurons, expressing both the pan-hippocampal ZBTB20 marker, typical of all neurons committed to hippocampal identity, and the KA1 marker, primarily expressed by pyramidal neurons in the CA3 subfield1 (see also Figure 9). Conversely, by using a high concentration of WNT3a (20 ng/mL) it is reported the specific induction of the DG hippocampal neurons phenotype.12,13
-
a.
Figure 6.
Representative brain organoids brightfield images and size at different stages of the culture protocols. Scale bars: 200μm
Figure 9.
Brain organoid's regional identity characterization
Representative confocal immunofluorescence images of sliced organoids at 32 days with hippocampal specification stained for pan-hippocampal (ZBTB20), CA3- (KA1) and CA2- (FZD9) hippocampal markers and sliced organoids at 32 days generated with the unguided protocol stained for choroid plexus (TTR, positive cells indicated by white arrows), cortical (FOXG1) and CA1- hippocampal (OCT6, positive cells indicated by white arrows) markers. Scale bars: 50μm.
Characterization of brain organoids by immunofluorescence:
Brain organoids’ fixing for immunofluorescence
Timing: 20 min
Fixing the brain organoids at critical time points allow to perform IF analysis and compare their development at different stages. Early organoids (before the differentiation phase) are very fragile so must be handled with care. On the contrary, late organoids, although being more compact, are smaller. Fixing brain organoids at different stage of the protocol, e.g., at 7, 14, 21 and 32 days, allows us to have a snap of the cellular organization at each phase of the culture as well as to follow the neuronal maturation. Any other desired time point can be selected to perform the analysis.
-
41.Collect the brain organoid by using a p1000 with cut tip, at any desirable time points.
-
a.Put the organoid in a well of a 24-well plate.
-
b.Aspirate and discard any medium residual.
-
c.Add 500 μL of fixing solution (see “materials and equipment” section for fixing solution recipe).
-
a.
-
42.
Incubate at 20°C–22°C for 15 min.
CRITICAL: longer periods of incubation will determine the deterioration of the 3D structures.
-
43.
Aspirate and remove the fixing solution and replace with 500 μL of PBS 1×.
-
44.
Aspirate and remove the PBS 1× with any residual of fixing solution and replace with 500 μL of PBS 1×.
CRITICAL: To ease this passage, put a black sheet under the plate in order to better visualize the samples and tilt the plate to let the brain organoids lay on a side.
-
45.
Repeat step 44.
-
46.
Store the fixed brain organoids in PBS 1× at 4°C for no longer than 3 weeks.
Note: for best results proceed to step 47 (for immunofluorescence on sliced brain organoids) or step 68 (for immunofluorescence on whole mount brain organoids) within a week.
Cryosectioning of brain organoids for immunofluorescence
Timing: 30 min
In this section we describe the steps to obtain 30 μm organoid’s slices for further immunostainings. Thinner slices are very likely to break during the cutting. Given their smaller dimensions compared to human brain organoids, be careful not to lose the organoids during the various steps. Early organoids (before differentiation phase), although having bigger size, are more fragile compared to the late ones. Refer to Figure 7 to visualize all the passages of the brain organoids’ cutting procedure.
CRITICAL: carefully avoid air bubbles throughout all the steps, since they will determine the breaking/loss of the brain organoid’s slices.
Note: for best results, it is important to maintain constant the working temperature of the cryostat (−20°C/−25°C) during all the steps. Keeping the room at cool temperature may help avoiding unwanted increased temperature of the instrument.
-
47.Create OCT stamps.
- a.
-
b.Keep the OCT stamps at −20°C.Note: OCT stamps can be prepared in advance.
-
48.
Once the OCT is solidified, define the zone where the brain organoid will be placed with a dark marker (Figure 7D). This step will ease the cutting process by making the organoid visible when it will be placed on the stamp.
- 49.
Note: Work at 20°C–22°C.
CRITICAL: minimize the volume of PBS transferred with the organoid. Troubleshooting 3.
-
50.
Pick up the brain organoid from the OCT drop using a needle and place it on the marked zone of the OCT stamp (Figure 7N).
CRITICAL: carefully pick up the organoid with the back part of the needle (Figures 7K‒7M), since the organoid gets stuck into the gauge. Alternatively, use a cut pipette tip but be careful not to get the organoid stuck in it.
-
51.
Place the stamp containing the brain organoid into the cryostat to allow the sample freezing.
-
52.
Prepare cryostat chucks, covering them with a thin layer of OCT (Figures 7O and 7P).
-
53.
When the sample is frozen, cover the brain organoid into the stamp with additional OCT. Let it freeze.
-
54.
Once the brain organoid is frozen, place the stamp vertically on the cryostat chunk and use some liquid OCT to allow it to stick to the chunk (Figure 7Q).
-
55.
Cover the structure with additional OCT to obtain a resistant structure that will not detach from the chunk while cutting (Figure 7R).
-
56.
When the structure is frozen, start cutting at 30μm (Figures 7S–7V).
-
57.
Check the integrity of the brain organoids slices under microscope (Figure 7W) and dark mark the glass slice in correspondence of organoid’s slices to ease their visualization.
Pause point: slices can be eventually stored at −20°C until use or directly used (step 59).
Figure 7.
Steps of organoids’ cutting for immunofluorescence analysis
Immunofluorescence on sliced brain organoids
Timing: 8 h
In this section we report the protocol for immunofluorescence analysis on sliced brain organoids, that allow to visualize the spatial distribution of the cells composing the organoids and to characterize their phenotype, as well as their developmental stage. By immunofluorescence analysis at defined time point i.e., 7, 14, 21 and 32 days it is possible to follow the progression of cellular organization and maturation within the brain organoids. The list of antibodies used to characterize the mouse brain organoids is reported in Table 1. To characterize the brain organoid development, we use Vimentin and SOX2 antibodies for neural stem cells14,15,16; Ki67 marker for proliferative cells17,18; DCX, B3Tubulin and MAP2 markers for neuronal precursors cells, immature and mature neurons,19,20,21,22,23 respectively; GFAP marker is used for glial cell identification.24,25 Representative images of immunofluorescence analysis performed on sliced brain organoids at different time point are reported in Figure 8.
Table 1.
List of primary antibodies and related target used to perform immunofluorescence analysis on brain organoids
| Antibody | Target |
|---|---|
| Brain organoid’s developmental characterization | |
| SOX2 (R&D System, # AF2018) | Neural stem cells |
| VIMENTIN (Millipore, # AB5733) | Neural stem cells |
| KI67 (Abcam, # ab16667) | Proliferative cells |
| DCX (Cell signaling technology, #4604) | Neuronal precursors cells |
| B3 TUBULIN (Promega, #G7121) | Immature neurons |
| MAP2 (Sigma-Aldrich, #M1406) | Mature neurons |
| GFAP (Abcam, #ab53554) | Astrocytes |
| SYNAPTOPHYSIN (Synaptic System, #101 004) | Synaptic puntae |
| ZBTB20 (Genetex, #GTX121616) | Pan-hippocampal cells |
| KA1 (Abcam, #ab67404) | CA3-hippocampal cells |
| PSD95 (Millipore, #MAB1596) | Post-synaptic terminals of the excitatory synapses |
| GEPHYRIN (Synaptic System, #147 011) | Post-synaptic terminals of the inhibitory synapses |
| VGAT (Synaptic System, #131 004) | Pre-synaptic terminals of the inhibitory synapses |
| VGLUT (Synaptic System, #135 303) | Pre-synaptic terminals of the excitatory synapses |
| GAD65/67 (Santa Cruz biotechnology, #sc-365180) | Inhibitory neurons |
| NMDA (Santa Cruz biotechnology, #sc-365597) | Excitatory neurons |
| GLSYN (Santa Cruz biotechnology, #sc-74430) | Glutamine synthetase produced by astrocytes |
| Brain organoid’s regional identity characterization | |
| GSX2 (Genetex, #GTX129390) | Ganglionic eminences |
| FOXG1 (Abcam, #ab18259) | Cortical neurons |
| NKX2.1 (Genetex, #GTX34907) | Ganglionic eminences |
| PREALBUMIN (TTR) (Genetex, #GTX85112) | Choroid plexus |
| FRIZZLED 9 (FZD9) (Genetex, #GTX71581) | CA2-hippocampal cells |
| OCT6 (Abcam, #ab272925) | CA1-hippocampal cells |
| PROX1 (Abcam, #ab101851) | Dentate gyrus cells |
Figure 8.
Representative confocal images of immunofluorescence analysis performed on sliced organoids generated with the unguided protocol at different stages of the culture protocol to evaluate the cellular organization and maturation. Scale bars: 50μm
Immunofluorescence analysis on sliced organoids can also give information about specific brain regional identity. Indeed, specific brain regional markers can be used to highlight the presence of hippocampal (i.e., ZBTB20, Ka1, OCT6 and FZD9)12,13,26 rather than cortical (i.e., FOXG1) cells within brain organoids. Pre-albumin (TTR) and NKX2.1 or GSX2 can be used to detect cells with choroid plexus and ganglionic eminences signature,26 respectively. Representative images of sliced organoids stained with brain regional identity markers are reported in Figure 9.
Note: Check the integrity of the brain organoids’ slices at the microscope. Perform immunofluorescence staining only on well-preserved slices.
CRITICAL: be careful throughout all the steps, brain organoids’ slices may detach from the slide. To prevent this, we recommend to gently apply drop by drop each solution near the slices avoiding the direct contact with the organoid’s slice.
-
58.
Thaw the cryosectioned slices at 20°C–22°C.
-
59.
Add 200 μL of PBS 1× per slice to hydrate it.
-
60.
Remove PBS 1× by gently drain the solution.
-
61.
Incubate brain organoids’ slices at 20°C–22°C for 1 h with 200 μL of blocking solution per slice.
Note: create a humidified chamber to prevent the evaporation of solutions.
CRITICAL: use 0.25% Triton-X for cytosolic markers and 0.5% Triton-X for nuclear markers.
-
62.
Drain the blocking solution and incubate with appropriate concentration of primary antibodies in 200 μL of blocking solution per slice for 2 h at 20°C–22°C.
Note: the primary antibodies validated on mouse brain organoids and used to characterize the organoids during development are reported in the key resources table and in Table 1. Please refer to Ciarpella et al.1 for further details. Any other antibodies against protein of interest could be used. Refer to related datasheet to check the applicability.
-
63.
Drain the primary antibodies solution and wash 6 times 5 min with 200 μL of blocking solution
-
64.
Drain the last blocking solution used for washing and incubate with secondary antibodies in 200 μL of blocking solution per slice for 1.5 h at 20°C–22°C in dark.
Note: the secondary antibodies validated on mouse brain organoids are reported in the key resources table. Please refer to Ciarpella et al.1 for further details. Any other secondary antibodies could be used. Refer to related datasheet to check the applicability.
-
65.
Drain the secondary antibodies solution and wash 3 times 5 min with 200 μL of blocking solution.
-
66.
Drain the last blocking solution used for washing and wash 3 times 5 min with 200 μL of PBS 1×.
-
67.At this step, nuclear staining is performed:
-
a.drain the PBS 1× and incubate each slice with TOPRO-3 in 200 μL of PBS 1× per slice for 10 min at 20°C–22°C in dark.Note: Any other nuclear staining could be performed, also accordingly to the wavelength of the laser’s microscope used.
-
b.Drain the nuclear staining solution and wash once with 200 μL of PBS 1×.
-
c.Drain the PBS 1× and close slides with 2 drops of DABCO and the glass coverslip, avoiding the formation of DABCO bubbles. Seal the glass coverslip on the slide with transparent nail polish.Alternatives: ready-to-use commercial Mountant reagent could be used rather than DABCO.Note: to ensure the optimal imaging, we usually acquire by confocal microscope the stained slices the following day.
-
a.
Immunofluorescence on whole mount brain organoids
Timing: 48 h
In this section we report the protocol to perform immunofluorescence on whole mount brain organoids. Preserving the 3D structure, this protocol allows us to analyze the network formation and gives information about the 3D spatial architecture and morphology of the organoids. To perform whole mount immunofluorescence, brain organoids must be previously fixed in Fixing solution (see steps 41–46).
-
68.
Put the fixed organoid in a well of a 96-well plate and incubate it with 80 μL of 0.5% blocking solution at 20°C–22°C for 1 h.
CRITICAL: always use 0.5% Triton-X when performing the whole mount IF.
-
69.
Carefully remove the blocking solution avoiding withdrawing the organoid and incubate with appropriate concentration of primary antibodies (see Table 1 and key resources table) in 80 μL of blocking solution per organoid at 4°C 12 h, on an orbital shaker (60 rpm).
-
70.Remove the primary antibodies solution:
-
a.wash 6 times 5 min with 80 μL of blocking solution.
-
a.
-
71.
Remove the last blocking solution used for washing and incubate with secondary antibodies in 80 μL of blocking solution per organoid for 6 h, at 20°C–22°C, static, in dark.
-
72.Remove the secondary antibodies solution:
-
a.wash 3 times 5 min with 80 μL of blocking solution.
-
a.
-
73.
Replace the last blocking solution used for washing with PBS 1×, wash 3 times 5 min.
-
74.Perform the nuclear staining:
-
a.remove the PBS 1×.
-
b.incubate each organoid with TOPRO-3 in 80 μL of PBS 1× for 10 min at 20°C–22°C in dark.
-
a.
Alternatives: Any other nuclear staining could be performed, also accordingly to the wavelength of the laser’s microscope used.
-
75.
Remove the nuclear staining solution and wash once with 80 μL of PBS 1×.
-
76.
Create a parafilm chamber on the slide (see Figure 10).
CRITICAL: this step is important to preserve the 3D morphology of the brain organoids. The parafilm chamber should be as thick as the sample to prevent its mashing and to avoid any extra space between the organoid and coverslip.
-
77.
Place the organoid inside the chamber and close it with 1 drop of DABCO using a round coverslip.
CRITICAL: Do not break the organoid while moving it. Make sure the organoid does not move into the corners of the chamber while closing with DABCO. Avoid the formation of DABCO bubbles.
Alternatives: ready-to-use commercial Mountant reagent could be used rather than DABCO.
-
78.
Seal the glass coverslip above the chamber by applying transparent nail polish at the edge.
Note: Images acquisition should be performed as soon as possible, since organoids will either move inside the chamber or become auto-fluorescent as time passes by.
Figure 10.
Immunofluorescence on whole mount brain organoids
(A) Parafilm chamber for whole mount organoid immunostaining confocal microscope acquisition.
(B) Representative maximum intensity projections of the z-stack confocal images of the entire mouse brain organoid stained for the mature neuronal marker MAP2 at 32 days. Scale bar: 200μm.
(C) Representative maximum intensity projections of the z-stack confocal images of a region of the entire mouse brain organoid stained for the mature neuronal marker MAP2, the astrocytes marker GFAP and the neuronal progenitors’ cells marker DCX. Scale bar: 50μm.
Functional assessment of brain organoids’ by using Fluo-4 calcium assay
Timing: 5 h
Functional assessment of the brain organoids’ spontaneous activity provides information about the maturation of the forming organoids.27,28 Fluo-4 assay on whole mount organoids allows detecting cellular activity preserving the organoids’ structure, rather than dissociating or slicing them as performed in several published protocols.29,30,31,32 This is a rapid assay that could be easily used to discriminate the effect of specific drugs on the cellular maturation and functionality.
Note: Fluo-4 is not exclusive for neuronal activity, but also for glial one. To exclusively examinate the neuronal activity, GcAMP6 lentiviral construct under the synaptophysin promoter can be used.33
-
79.
Wash the brain organoids with HEPES 0.3 M pH 7.3 for 10 min.
-
80.
Incubate organoids for 3 h at 37°C with a solution of 50% Fluo4 + 50% PSS (100 μL for each organoid).
CRITICAL: work in sterile conditions, protected from light.
-
81.
Wash organoids three times with culture medium.
-
82.
Incubate organoids for 30 min at 37°C with appropriate culture medium.
Note: this step ensures that all the AM-group of the Fluo4 are de-esterification so that the fluorescence signal is constant during acquisition.
-
83.
Perform time-lapse analysis using the Nikon microscope, at 37°C and 5% CO2. Troubleshooting 4.
Alternatives: other fluorescence, confocal, two-photon microscopes equipped with laser and/or lamp with the desired wavelength can be used to visualize the Fluo4. A microscope equipped with a temperature and gas controller is highly recommended. Keeping the temperature and CO2 constant during acquisition help to avoid Ca2+ fluctuation artefacts.
-
84.Analyze the generated videos, for a qualitative indication of the Ca2+ flux, using ImageJ software:
-
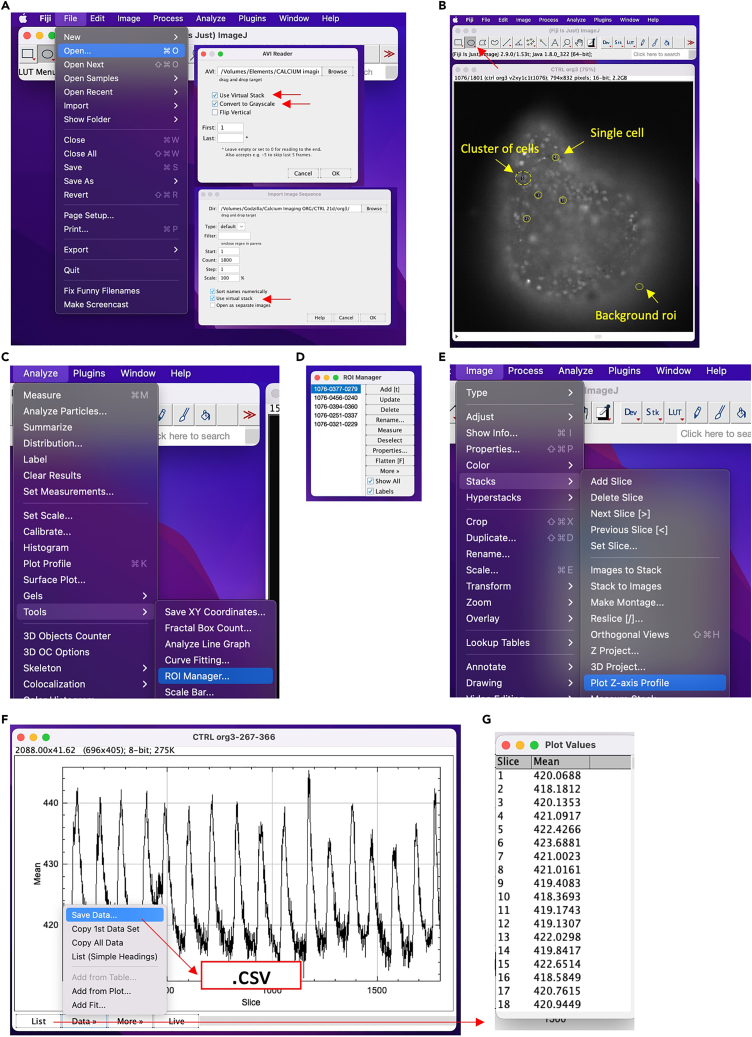
a.Open ImageJ software, then load the time-lapse images sequence: File > open.Note: an "Import image sequence” tab will be open. Be sure to select the “use virtual stack” option. ImageJ also supports file .avi. An “AVI reader” tab will be open and “use virtual stack” should be clicked. We recommend working in greyscale. (Figure 11A)
-
b.Draw a sufficient number of ROI for each organoid, related to a single cell or small group of cells: Oval/Rectangular selection > Analyze > Tools > ROI manager > add (Figures 11B–11D).Note: draw a ROI in a background zone for further quantitative analysis.
-
c.Select one ROI by one from the ROI manager: Image > Stacks > Plot Z-axis Profile (Figure 11E).
-
d.A graph showing the Ca2+ fluctuations is now open (Figure 11F) and, if needed, it could be saved.
-
e.Save the values related to the graph for further analysis: Data > Save data > (.xls/.csv).Note: it is also possible to save the list of values, to be processed with any other analysis software: List > Plot Values (Figures 11F and 11G).Alternatives: different software or custom-made calcium analyzer could be used to quantify Ca2+ flux and cellular activity. We report here a fast analysis to qualitatively analyze the Fluo4 assay output.
-
a.
Figure 11.
Step-by-step of qualitative calcium imaging analysis using ImageJ software
DNA extraction from brain organoids
Timing: 4 h
Fully mature neurons display high energy consumption to sustain their active metabolism and thus, neuronal differentiation implies a switch from glycolytic to oxidative metabolism.34,35,36,37,38,39 Characterizing the metabolic profile of organoids allows us to acquire information about brain organoids’ development: early organoids rely mainly on glycolysis, while late organoids activate oxidative phosphorylation hence, lower mtDNA copy number and mitochondrial subunits amount are expected in early differentiated compared to fully mature organoids. The DNA extracted with this optimized protocol can be used to evaluate the mtDNA copy number, a good indicator of the amount of mitochondrial mass, by following the method reported in.40
-
85.
Put single organoids in a 1.5 mL Eppendorf, at any time point considered.
-
86.
Wash organoids with sterile PBS 1×.
-
87.
Spin quickly.
Note: step 87 can be skipped for early organoids.
-
88.
Carefully remove the supernatant.
CRITICAL: organoids must be dry after this step.
Pause point: organoids can be eventually stored at −20°C or directly used (step 90).
-
89.
On the day of the extraction thaw your samples and keep them on ice.
-
90.
Add 250 μL of lysis buffer plus 2.5 μL of Proteinase K (10 mg/mL) inside the Eppendorf with the organoid and lysate the sample, avoiding bubbles.
-
91.
Incubate at 55°C in a thermoblock for 2–2.5 h and vortex occasionally.
-
92.
At the end of the incubation time, vortex the Eppendorf and centrifuge for 10 min at maximum speed, 20°C–22°C.
-
93.
The pellet contains cellular debris. Take the supernatant and put it in a new Eppendorf.
-
94.
Add 250 μL (1 volume) of isopropyl alcohol and 1 μL of glycogen (it helps visualizing the DNA).
CRITICAL: glycogen concentration cannot be over 4 μg/μL, otherwise it may inhibit PCR reaction.
Note: if DNA clump is not visible, put the Eppendorf at −80°C for about 30 min, it helps the glycogen precipitate the DNA.
-
95.
Centrifuge at 17.530 g for 30 min at 20°C–22°C.
-
96.
Wash the DNA pellet with 50 μL of EtOH 70%.
-
97.
Air-dry the pellet.
-
98.
Dissolve the pellet into 30 or 50 μL for late or early organoids, respectively of TE buffer pH 7.5.
-
99.
Quantify the DNA concentration using the Nanodrop.
Pause point: DNA may be stored at −20°C until use or directly used (step 100).
Real-time PCR for mitochondrial DNA
Timing: 4 h
Evaluation of the mtDNA copy number can be a good indicator of the amount of mitochondrial mass. We report here the steps to perform the PCR reaction for the analysis of mtDNA/nDNA ratio in mouse cells using a SYBR green assay. It will allow to estimate the mtDNA copy number, by comparing the amount of mitochondrial versus nuclear DNA.40
Note: In a single 96 well plate, two different amplification reactions will be run: one for the genomic DNA, and one for the mitochondrial DNA; so ideally the plate will be divided in two sections. Each sample will run in three technical replicates for each amplification reaction. In the following steps, solutions amount is calculated for one sample in three technical replicates.
-
100.
Dilute the DNA samples at a concentration of 5 ng/μL.
-
101.Prepare two separate PCR working solution (48 μL total volume each; see “materials and equipment” section for PCR working solution receipt):
-
a.Genomic PCR working solution containing forward and reverse primers for the nuclear gene HK2 (hexokinase 2).
-
b.Mitochondrial PCR working solution containing forward and reverse primers for the mitochondrial-encoded gene ND1.
-
a.
-
102.
Add 12 μL of DNA sample to each PCR working solution (genomic and mitochondrial).
-
103.
Load the plate with 20 μL/well of the solutions obtained in step 102.
-
104.
Seal the plate with the appropriate coverslip.
-
105.
Set the software and run the PCR with the following settings for 45 cycles:
Hold Stage: 50°C for 2 min followed by 95°C for 5 min.
PCR Stage: 95°C for 5 s followed by 60°C for 1 min.
Melt curve Stage: 95°C for 5 s, 66°C for 1 min, 97°C for 1 s.
Note: we use the Real Time QS3 system (Thermofisher) equipped with QuantStudio™ Design & Analysis Software v1.5.1.
-
106.
Calculate the number of copies of mtDNA with the following formulas40 :
Protein isolation and analysis of brain organoids
Timing: 2 h
These steps allow the extraction of the protein content of the brain organoids. Organoids change in the morphology, dimension and cellular density during the in vitro culturing protocol. We optimized the number of organoids to be pooled together at each time point and the volume of solutions in order to have a reasonable yield of material. This resulted in pools of different number of organoids depending on the stage of maturation.
-
107.
Collect pools of 3 organoids (for early organoids: day 7 and day 14) or 5 organoids (late organoids: day 21 and day 32) in 500 μL Eppendorf.
-
108.
Wash organoids with sterile PBS 1×.
-
109.
Spin quickly.
Note: for early organoids you can avoid this step.
-
110.
Remove the supernatant.
CRITICAL: organoids must be dry after this step.
Pause point: organoids may be stored at −80°C until use or directly used (step 111).
-
111.
Resuspend the pools of organoids in 25 or 13 μL of Complete RIPA buffer for early and late organoid respectively, dissociate by pipetting until the organoids are not visible.
Note: early organoids are quite easy to dissociate, since they are not very compact; the late ones may require a bit more effort to completely dissociate.
-
112.
Vortex and keep on ice for 30 min. Vortex every 10 min.
-
113.
Add 1 μL of Benzonase per sample and incubate at 20°C–22°C for 30 min.
-
114.
Centrifuge at 10000 g for 15 min at 4°C.
-
115.
Transfer the supernatant, that contains the extracted proteins, to a new Eppendorf.
-
116.
Quantify extracted proteins with BCA assay kit following manufacture instructions (https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0011430_Pierce_BCA_Protein_Asy_UG.pdf). Troubleshooting 5.
Neuronal differentiation requires a switch from glycolytic to oxidative metabolism35 and an increase of OXPHOS subunits and mitochondrial proteins is expected during organoid maturation. Extracted proteins are used to perform Western Blot analysis to detect and quantify the mitochondrial proteins and OXPHOS system quantity. The voltage-dependent anion-selective channel 1 (VDAC 1) protein, a mitochondrial channel of the outer mitochondrial membrane, is used as an indicator of mitochondrial mass while the amount of mitochondrial respiratory chain complexes I-V is given by the OXPHOS cocktail antibody. We usually express the mitochondrial markers as fold-change of the nuclear marker Lamin B, rather than to the total protein content per lane. Indeed, given the heterogeneity of cell composition of brain organoids across the developmental stages, equal amounts of protein/organoid pool at different maturation phases may not reflect the cell number. Specific antibodies can be used for any other proteins of interest.
Expected outcomes
We established a reliable and reproducible three-phase protocol to generate functional brain organoids from mouse embryonic (E14.5) neural stem cells (NSCs) in 32 days, which displayed consistent size and functional features. The steps include a fine-tuning of media composition and application time, allowing the progressive neuronal cells maturation and organization into a 3D-brain like tissue. By using this step-by-step protocol, a 95% rate of successful brain organoid formation and maintenance along the in vitro culture timing (32 days) with a 5% rate of brain organoid disintegration after 14 days or no aggregation within the first 48 h is expected. The cellular organization and the progressive neuronal differentiation of the generate murine brain organoids can be observed by several techniques such as immunofluorescence analysis, western blot and calcium imaging. Extensive data about brain organoids characterization resulting from these methodologies are reported in Ciarpella et al.1
Mouse brain organoid generated by using this protocol allows for insights into neuronal development as well as disease modeling and drug screening. It represents a more cost-effective and less-time consuming alternative to human brain organoid models. Indeed, even if human brain organoids are a powerful technology for translational potential and precision medicine, mouse derived organoids represent a useful supportive tool to optimize protocols for human-derived brain organoids and to perform large-scale drug discovery studies before validation on human-derived models. Due to the possibility of obtaining genetically mutant phenotype samples, mouse organoids can also serve as systems to study the pathophysiology of different brain disorders, reducing the usage of animals.
Limitations
This work reports the generation of functional brain organoids starting from mouse embryonic neural stem cells in a time- and cost- efficient manner. However, there are some technical limitations to the protocol. Due to the small dimension of mouse brain organoids, a functional characterization of the neuronal network by technique such as multielectrode array (MEA) systems is hard to be addressed. Small organoids require small and high-density electrodes in order to detect neuronal activity. Unless the small dimension, the lack of vascular systems that ensure proper supplies of nutrition and gas exchange could lead to the formation of a necrotic core in the later stage of differentiation, which may affect the final readouts. To the best of our knowledge, mouse brain organoid model is populated by different cellular type such as neuronal precursors cells, immature and mature neurons, astrocytes and, at least in part during the mature stage, stem cells. Adding cellular components such as microglia or oligodendrocytes may represent an improvement of the proposed protocol, which could benefit in terms of functional maturation and cellular complexity.
Troubleshooting
Problem 1
More than one brain organoid is forming in a single well (related to Step 35a and 38a).
Potential solution
Following Accutase treatment ensure that each neurosphere is correctly dissociated in single cells. If some cellular aggregates persist after splitting, the cell seeding at day 0 will be not homogeneous. The coexistence of single cells and cellular aggregates will result in the formation of multiple cellular aggregates that could led to an incorrect organoid formation. Check the 24-well plate at the microscope after seeding and exclude from the analysis the wells containing cellular aggregate.
Problem 2
Brain organoids are not forming within 48 h (related to Step 35b and 38b).
Potential solution
We recommend seeding the cells in the center of the well and to not spread them around the well to facilitate cells aggregation and organoid formation. To do this, place the pipette’s tip at the center of the well and keep the position while gently pipetting up and down the cell suspension solution. Seed the precise number of cells in each well of a 24-wells plate by counting them accurately and ensuring homogeneous pipetting by mixing up and down the cells in the falcon tube before each seeding in 24-wells. Brain organoid’s formation could also be impacted by cells splitting. Accutase treatment should be performed as gently as possible. Ensure to not proceed for more than 5 min in order to prevent cell death. Avoid seeding cellular suspension if the viability is less than 90%.
Problem 3
Brain organoids collapse or break during cryostat sectioning and the resulted slices cannot be further use for IF analysis (related to Step 49).
Potential solution
When the organoid is transferred from PBS to the OCT drop, ensure to pick up the organoid with the less PBS 1× solution possible. To favor the removal of PBS 1× excess, subsequently transfer the organoid from an OCT drop to another one until a small amount of PBS 1× is present. Reducing the cutting speed could also help to keep the slice intact during the procedure. If the problem persists, it is possible to increment the slice thickness.
Problem 4
Brain organoids doesn’t reveal any calcium activity (related to Step 83).
Potential solution
Prolongated Fluo4-am dye incubation could result in saturated signal. When the signal is too high, it is difficult to detect intensity variation in the fluorescence. The incubation time could depend on organoid dimension: small organoids could benefit from an incubation of 2 h. Prolongated Fluo4-am dye incubation could also lead to cell dead: brightest cells are probably suffering. Avoid including these cells in the subsequent analysis. The best activity signal is expected to be at 21/32 days in culture. Little activity is observed at day 7, probably coming from glial cells, while an increment is observed starting from day 14.
Problem 5
Low or insufficient yield of protein amount is obtained after protein extraction procedure (related to Step 116).
Potential solution
Incomplete or not-efficient organoid lysis could lead to low protein amount. Ensure to completely dry the organoid before adding the Complete RIPA buffer. Due to the small volume of buffer used, any residual of PBS will dilute it and reduce the efficiency of the organoid lysis. Avoiding bubble formation during lysis could favor the tissue-like visualization and thus can help to evaluate when to stop the lysis process. If the amount of protein obtained is still not sufficient for further desired analysis, the number of organoids to be pooled together could be incremented.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ilaria Decimo (ilaria.decimo@univr.it)
Materials availability
This study did not generate any unique reagents.
Acknowledgments
We acknowledge Dr. Sissi Dolci for the initial help in the setting up the protocol and Andrea Borioli; “Centro Interdipartimentale di Servizi per la Ricerca che utilizza Animali da Laboratorio” – C.I.R.S.A.L. and “Centro Piattaforme Tecnologiche” – CPT (University of Verona) are acknowledged for services and support. European Union project FETPROACT-2018-2020 HERMES (grant number 824164), Fondazione Telethon–Italy (Grant Number GSP20004_PAsMCT8006) are acknowledged for the support on research provided to I.D. Work is supported by #NEXTGENERATIONEU (NGEU) and funded by the Ministry of University and Research (MUR), National Recovery and Resilience Plan (NRRP), project MNESYS (PE0000006) – A Multiscale integrated approach to the study of the nervous system in health and disease (DN. 1553 11.10.2022). Figure 1A of this paper was created with BioRender.com.
Author contributions
F.C. designed, implemented, and wrote the protocol; R.G.Z., A.C., and G.P. designed, implemented the protocol and revised the manuscript; M.D.C. and E.B. participated in method development and revised the manuscript. I.D. conceived, designed, supervised the study, and revised the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2023.102413.
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Ciarpella F., Zamfir R.G., Campanelli A., Ren E., Pedrotti G., Bottani E., Borioli A., Caron D., Di Chio M., Dolci S., et al. Murine cerebral organoids develop network of functional neurons and hippocampal brain region identity. iScience. 2021;24:103438. doi: 10.1016/j.isci.2021.103438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Decimo I., Dolci S., Panuccio G., Riva M., Fumagalli G., Bifari F. Meninges: a widespread niche of neural progenitors for the brain. Neuroscientist. 2021;27:506–528. doi: 10.1177/1073858420954826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pino A., Fumagalli G., Bifari F., Decimo I. New neurons in adult brain: distribution, molecular mechanisms and therapies. Biochem. Pharmacol. 2017;141:4–22. doi: 10.1016/j.bcp.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Fuentealba L.C., Rompani S.B., Parraguez J.I., Obernier K., Romero R., Cepko C.L., Alvarez-Buylla A. Embryonic origin of postnatal neural stem cells. Cell. 2015;161:1644–1655. doi: 10.1016/j.cell.2015.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martynoga B., Drechsel D., Guillemot F. Molecular control of neurogenesis: a view from the mammalian cerebral cortex. Cold Spring Harb. Perspect. Biol. 2012;4:a008359. doi: 10.1101/cshperspect.a008359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeadon J. 2014. 6 Steps for Setting up Timed Pregnant Mice.https://www.jax.org/news-and-insights/jax-blog/2014/september/six-steps-for-setting-up-timed-pregnant-mice [Google Scholar]
- 7.Roustan A., Perrin J., Berthelot-Ricou A., Lopez E., Botta A., Courbiere B. Evaluating methods of mouse euthanasia on the oocyte quality: cervical dislocation versus isoflurane inhalation. Lab. Anim. 2012;46:167–169. doi: 10.1258/la.2012.011115. [DOI] [PubMed] [Google Scholar]
- 8.Wuri L., Agca C., Agca Y. Euthanasia via CO2 inhalation causes premature cortical granule exocytosis in mouse oocytes and influences in vitro fertilization and embryo development. Mol. Reprod. Dev. 2019;86:825–834. doi: 10.1002/mrd.23167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang B.S., Lowenstein D.H. N. Engl. J. Med. 2003;349:1257–1266. doi: 10.1056/NEJMra022308. [DOI] [PubMed] [Google Scholar]
- 10.Dhikav V., Anand K. Hippocampal atrophy may be a predictor of seizures in Alzheimer's disease. Med. Hypotheses. 2007;69:234–235. doi: 10.1016/j.mehy.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 11.Lie D.C., Colamarino S.A., Song H.J., Désiré L., Mira H., Consiglio A., Lein E.S., Jessberger S., Lansford H., Dearie A.R., Gage F.H. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- 12.Sakaguchi H., Kadoshima T., Soen M., Narii N., Ishida Y., Ohgushi M., Takahashi J., Eiraku M., Sasai Y. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 2015;6:8896. doi: 10.1038/ncomms9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarkar A., Mei A., Paquola A.C.M., Stern S., Bardy C., Klug J.R., Kim S., Neshat N., Kim H.J., Ku M., et al. Efficient generation of CA3 neurons from human pluripotent stem cells enables modeling of hippocampal connectivity in vitro. Cell Stem Cell. 2018;22:684–697.e9. doi: 10.1016/j.stem.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis P., Fagan B.M., Magness S.T., Hutton S., Taranova O., Hayashi S., McMahon A., Rao M., Pevny L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev. Neurosci. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- 15.Pevny L.H., Nicolis S.K. Sox2 roles in neural stem cells. Int. J. Biochem. Cell Biol. 2010;42:421–424. doi: 10.1016/j.biocel.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 16.Vinci L., Ravarino A., Fanos V., Naccarato A.G., Senes G., Gerosa C., Bevilacqua G., Faa G., Ambu R. Immunohistochemical markers of neural progenitor cells in the early embryonic human cerebral cortex. Eur. J. Histochem. 2016;60:2563. doi: 10.4081/ejh.2016.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller I., Min M., Yang C., Tian C., Gookin S., Carter D., Spencer S.L. Ki67 is a graded rather than a binary marker of proliferation versus quiescence. Cell Rep. 2018;24:1105–1112.e5. doi: 10.1016/j.celrep.2018.06.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun X., Kaufman P.D. Ki-67: more than a proliferation marker. Chromosoma. 2018;127:175–186. doi: 10.1007/s00412-018-0659-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breuss M.W., Leca I., Gstrein T., Hansen A.H., Keays D.A. Tubulins and brain development - the origins of functional specification. Mol. Cell. Neurosci. 2017;84:58–67. doi: 10.1016/j.mcn.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Brown J.P., Couillard-Després S., Cooper-Kuhn C.M., Winkler J., Aigner L., Kuhn H.G. Transient expression of doublecortin during adult neurogenesis. J. Comp. Neurol. 2003;467:1–10. doi: 10.1002/cne.10874. [DOI] [PubMed] [Google Scholar]
- 21.Gleeson J.G., Lin P.T., Flanagan L.A., Walsh C.A. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- 22.Johnson G.V., Jope R.S. The role of microtubule-associated protein 2 (MAP-2) in neuronal growth, plasticity, and degeneration. J. Neurosci. Res. 1992;33:505–512. doi: 10.1002/jnr.490330402. [DOI] [PubMed] [Google Scholar]
- 23.Luzzati F., Bonfanti L., Fasolo A., Peretto P. DCX and PSA-NCAM expression identifies a population of neurons preferentially distributed in associative areas of different pallial derivatives and vertebrate species. Cereb. Cortex. 2009;19:1028–1041. doi: 10.1093/cercor/bhn145. [DOI] [PubMed] [Google Scholar]
- 24.Jurga A.M., Paleczna M., Kadluczka J., Kuter K.Z. Beyond the GFAP-astrocyte protein markers in the brain. Biomolecules. 2021;11:1361. doi: 10.3390/biom11091361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z., Ma Z., Zou W., Guo H., Liu M., Ma Y., Zhang L. The appropriate marker for astrocytes: comparing the distribution and expression of three astrocytic markers in different mouse cerebral regions. BioMed Res. Int. 2019;2019:9605265. doi: 10.1155/2019/9605265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Renner M., Lancaster M.A., Bian S., Choi H., Ku T., Peer A., Chung K., Knoblich J.A. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 2017;36:1316–1329. doi: 10.15252/embj.201694700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenberg S.S., Spitzer N.C. Calcium signaling in neuronal development. Cold Spring Harb. Perspect. Biol. 2011;3:a004259. doi: 10.1101/cshperspect.a004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakaguchi H., Ozaki Y., Ashida T., Matsubara T., Oishi N., Kihara S., Takahashi J. Self-organized synchronous calcium transients in a cultured human neural network derived from cerebral organoids. Stem Cell Rep. 2019;13:458–473. doi: 10.1016/j.stemcr.2019.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li R., Sun L., Fang A., Li P., Wu Q., Wang X. Recapitulating cortical development with organoid culture in vitro and modeling abnormal spindle-like (ASPM related primary) microcephaly disease. Protein Cell. 2017;8:823–833. doi: 10.1007/s13238-017-0479-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paşca A.M., Sloan S.A., Clarke L.E., Tian Y., Makinson C.D., Huber N., Kim C.H., Park J.Y., O'Rourke N.A., Nguyen K.D., et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C., et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi Y., Sun L., Wang M., Liu J., Zhong S., Li R., Li P., Guo L., Fang A., Chen R., et al. Vascularized human cortical organoids (vOrganoids) model cortical development in vivo. PLoS Biol. 2020;18:e3000705. doi: 10.1371/journal.pbio.3000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen T.W., Wardill T.J., Sun Y., Pulver S.R., Renninger S.L., Baohan A., Schreiter E.R., Kerr R.A., Orger M.B., Jayaraman V., et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beckervordersandforth R., Ebert B., Schäffner I., Moss J., Fiebig C., Shin J., Moore D.L., Ghosh L., Trinchero M.F., Stockburger C., et al. Role of mitochondrial metabolism in the control of early lineage progression and aging phenotypes in adult hippocampal neurogenesis. Neuron. 2017;93:560–573.e6. doi: 10.1016/j.neuron.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bifari F., Dolci S., Bottani E., Pino A., Di Chio M., Zorzin S., Ragni M., Zamfir R.G., Brunetti D., Bardelli D., et al. Complete neural stem cell (NSC) neuronal differentiation requires a branched chain amino acids-induced persistent metabolic shift towards energy metabolism. Pharmacol. Res. 2020;158:104863. doi: 10.1016/j.phrs.2020.104863. [DOI] [PubMed] [Google Scholar]
- 36.Inak G., Rybak-Wolf A., Lisowski P., Pentimalli T.M., Jüttner R., Glažar P., Uppal K., Bottani E., Brunetti D., Secker C., et al. Defective metabolic programming impairs early neuronal morphogenesis in neural cultures and an organoid model of Leigh syndrome. Nat. Commun. 2021;12:1929. doi: 10.1038/s41467-021-22117-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorenz C., Lesimple P., Bukowiecki R., Zink A., Inak G., Mlody B., Singh M., Semtner M., Mah N., Auré K., et al. Human iPSC-derived neural progenitors are an effective drug discovery model for neurological mtDNA disorders. Cell Stem Cell. 2017;20:659–674.e9. doi: 10.1016/j.stem.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 38.Martano G., Borroni E.M., Lopci E., Cattaneo M.G., Mattioli M., Bachi A., Decimo I., Bifari F. Metabolism of stem and progenitor cells: proper methods to answer specific questions. Front. Mol. Neurosci. 2019;12:151. doi: 10.3389/fnmol.2019.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng X., Boyer L., Jin M., Mertens J., Kim Y., Ma L., Ma L., Hamm M., Gage F.H., Hunter T. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Elife. 2016;5:e13374. doi: 10.7554/eLife.13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quiros P.M., Goyal A., Jha P., Auwerx J. Analysis of mtDNA/nDNA ratio in mice. Curr. Protoc. Mouse Biol. 2017;7:47–54. doi: 10.1002/cpmo.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.