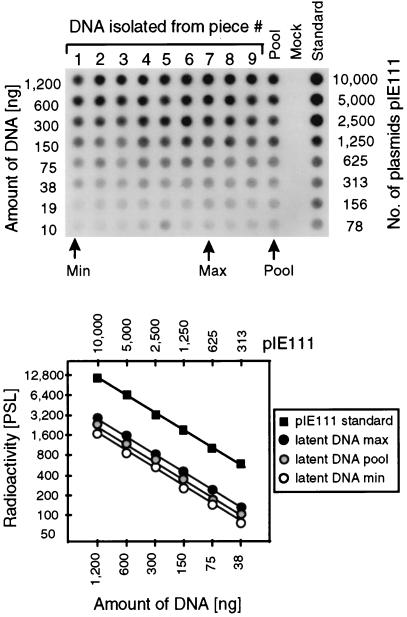
FIG. 4.
Determination of the viral DNA load during murine CMV latency in the lungs. In parallel with the isolation of the poly(A)+ RNA used for the transcriptional analyses shown in Fig. 3B and C, DNA was isolated from lung tissue pieces #1 through #9 derived from mouse LM1. The DNA from the individual pieces as well as a pool of the DNA from all nine pieces (Pool) were titrated and subjected to an ie1 exon 4-specific PCR. A negative control (Mock) was provided by DNA derived from uninfected lungs. As a standard for the quantitation, the mock DNA was supplemented with plasmid pIE111. (Top) Autoradiograph of the dot blot obtained after hybridization with a γ-32P-end-labeled internal oligonucleotide probe. (Bottom) Computed phosphorimaging results for the same blot. For the sake of clarity, the computations are depicted only for the pool, representing the average of all samples, as well as for the individual samples with the lowest load (Min) and the highest load (Max) among the nine samples tested. Log-log plots of radioactivity measured as phosphostimulated luminescence (PSL) units (ordinate) versus the amount of sample DNA (abscissa) are shown. The upper rule relates the amount of DNA to the number of plasmids in the pIE111 standard.