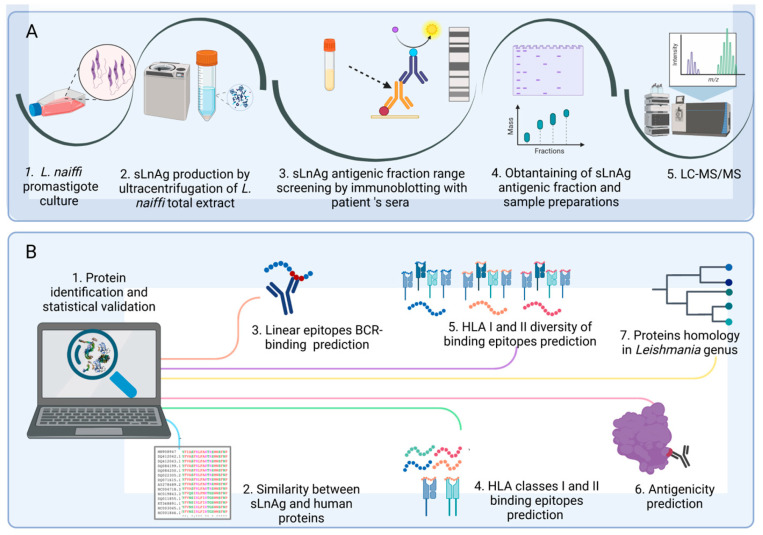
Figure 1.
Workflows of the proteomic and bioinformatic analysis. (A). Proteomic experiments. Cultured metacyclic promastigotes of L. naiffi originated total extract. sLnAg was obtained by ultracentrifugation of the total extract. Groups of sLnAg proteins were separated by SDS-PAGE and incubated with sera from cured CL patients. The molecular weight band with the highest frequency of antigen–antibody complexes was obtained in a new electrophoretic run under the same conditions as the previous one. Following FASP elution, the fractions were subjected to LC-MS/MS. (B). Bioinformatics analysis. The proteins of the antigenic fraction of sLnAg were identified from the mass spectra obtained. Subsequently, a protein-mining analysis was performed. We excluded proteins similar to human proteins and selected proteins with more BCR and HLA I and II binding epitopes, as well as greater diversity with respect to HLA allele binding. The selected proteins were analyzed for antigenicity using the Vaxijen algorithm and for conservation in the genus Leishmania, criteria to be considered as potential vaccine targets.