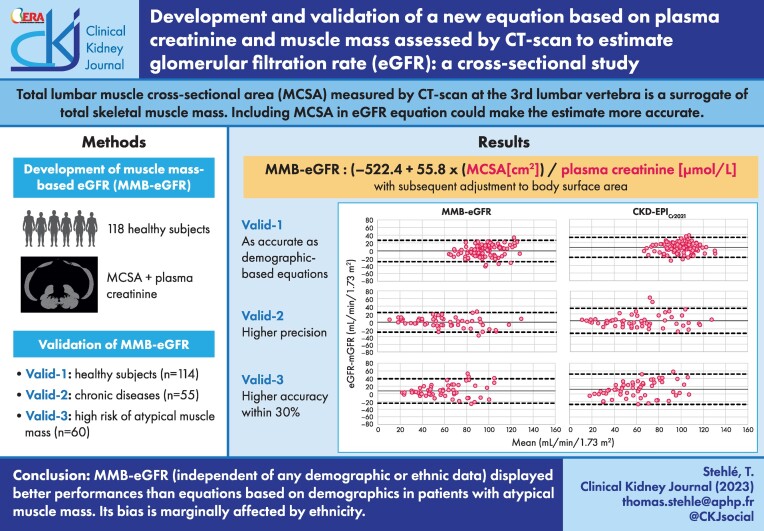
ABSTRACT
Background
Inter-individual variations of non-glomerular filtration rate (GFR) determinants of serum creatinine, such as muscle mass, account for the imperfect performance of estimated GFR (eGFR) equations. We aimed to develop an equation based on creatinine and total lumbar muscle cross-sectional area measured by unenhanced computed tomography scan at the third lumbar vertebra.
Methods
The muscle mass–based eGFR (MMB-eGFR) equation was developed in 118 kidney donor candidates (iohexol clearance) using linear regression. Validation cohorts included 114 healthy subjects from another center (51Cr-EDTA clearance, validation population 1), 55 patients with chronic diseases (iohexol, validation population 2), and 60 patients with highly discordant creatinine and cystatin C–based eGFR, thus presumed to have atypical non-GFR determinants of creatinine (51Cr-EDTA, validation population 3). Mean bias was the mean difference between eGFR and measured GFR, precision the standard deviation (SD) of the bias, and accuracy the percentage of eGFR values falling within 20% and 30% of measured GFR.
Results
In validation population 1, performance of MMB-eGFR was not different from those of CKD-EPICr2009 and CKD-EPICr2021. In validation population 2, MMB-eGFR was unbiased and displayed better precision than CKD-EPICr2009, CKD-EPICr2021 and EKFC (SD of the biases: 13.1 vs 16.5, 16.8 and 15.9 mL/min/1.73 m2). In validation population 3, MMB-eGFR had better precision and accuracy {accuracy within 30%: 75.0% [95% confidence interval (CI) 64.0–86.0] vs 51.5% (95% CI 39.0–64.3) for CKD-EPICr2009, 43.3% (95% CI 31.0–55.9) for CKD-EPICr2021, and 53.3% (95% CI 40.7–66.0) for EKFC}. Difference in bias between Black and white subjects was −2.1 mL/min/1.73 m2 (95% CI −7.2 to 3.0), vs −8.4 mL/min/1.73 m2 (95% CI −13.2 to −3.6) for CKD-EPICr2021.
Conclusion
MMB-eGFR displayed better performances than equations based on demographics, and could be applied to subjects of various ethnic backgrounds.
Keywords: creatinine, cystatin C, estimated glomerular filtration rate, muscle mass, sarcopenia, CT scan
Graphical Abstract
Graphical Abstract.
INTRODUCTION
In clinical practice, the estimation of the glomerular filtration rate (GFR) is usually derived from equations based on plasma or serum creatinine concentration, age and sex [1–3]. Ethnicity was also included in these equations, because at equal GFR value, African-American subjects have higher serum creatinine than non-Black subjects [1, 2]. Incorporating ethnicity into eGFR equations has become all the more controversial since race is a social construct and the race correction factor used in Afro-Americans could be seen as a form of inequality [4]. The US guidelines of September 2021 dropped the race variable [5], and recommended new Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equations omitting race [6]. Using creatinine-based eGFR equations is prompted by the simplicity, worldwide availability, standardization [7] and inexpensiveness of the creatinine assay. However, the accuracy of these equations is not optimum since the level of serum creatinine depends not only on GFR but also on non-GFR determinants, e.g, the amount of creatinine the body generates, the tubular secretion of creatinine and its extra-renal clearance [8]. Creatinine generation depends mainly on the body muscle mass and, to a lesser extent, on daily protein intake [9–11]. It has been hypothesized that African-American patients generate more creatinine than non-Black subjects due to differences in muscle mass [12, 13], but there are conflicting data [14, 15]. The demographic variables of creatinine-based eGFR equations, namely age and sex, do not help identify outliers, whose creatinine generation is atypical because of high or low muscle mass. These equations could gain more accuracy if they incorporated data on muscle mass. Such data can be non-invasively obtained from computed tomography (CT) imaging. The total lumbar muscle cross-sectional area (MCSA) taken at the middle of the third lumbar vertebra (L3) is currently considered as a surrogate marker of total body muscle mass [16–18]. It can be easily measured, from unenhanced CT scan images, with several medical imaging software, either automatically or semi-automatically [19–23]. For patients who have CT scans as part of their medical follow-up (especially patients with chronic diseases and thus at risk for sarcopenia), a GFR estimation equation that would include CT scan–based muscle data might offer the possibility of a more personalized eGFR, without additional cost.
The objectives of our study were first to evaluate the relationship between MCSA and creatinine generation, and secondly to develop and validate a new equation for GFR estimation by incorporating MCSA and plasma creatinine level.
MATERIALS AND METHODS
Study design
This is a cross-sectional study using data for four groups of patients who underwent abdominal CT scan within 6 months before or after the date their GFR was measured. The correlation between urinary creatinine excretion rate and MCSA was examined, then a GFR estimation equation was developed in kidney donor candidates (development population). The equation was validated in three external groups: kidney donor candidates from another center (validation population 1), patients with various chronic illnesses (validation population 2) and patients with highly discordant creatinine-based and cystatin C–based eGFR, thus presumed to have atypical non-GFR determinants of serum creatinine, including low or high muscle masses [24–26] (validation population 3). The Institutional Review Boards of the two centers approved this study: IRB-00011558/2021-103 and IRB-00006477/14-051, and the patients gave their informed consents.
Study populations
The development population included all living kidney donor candidates investigated in the Nephrology Department of Henri-Mondor Hospital (Creteil, Grand Paris Area, France) between July 2016 and June 2021. They underwent abdominopelvic CT scan to assess kidney morphology, and iohexol clearance to measure GFR (300 mg/L Omnipaque; GE Healthcare). Validation population 1 included all kidney donor candidates evaluated at Bichat Hospital (Paris, France), between April 2013 and December 2018, for whom both 51Cr-EDTA clearance (GE Healthcare) and abdominal CT scan results were available. Validation population 2 included all patients, except kidney donor candidates, who underwent iohexol clearance at Henri-Mondor Hospital and had abdominal CT scan within 6 months of GFR measurement over the same period as the development population. Validation population 3 included all patients from Bichat Hospital, except kidney donor candidates, who underwent CT scan within 6 months of having their GFR measured over the same period as validation population 1, and for whom CKD-EPICr2009 and CKD-EPICys were highly discordant, i.e, (CKD-EPICr2009 – CKD-EPICys)/mean was either >30% or <−30%. The only non-inclusion criteria was limb amputation, which might incur inaccurate estimation of muscle mass upon using MCSA.
Clinical and biological data
Sex, age, ethnicity, weight, height and medical history were collected from medical records.
Creatinine and cystatin C investigations are depicted in the Supplementary data.
GFR measurement and determining creatinine excretion rate
GFR was measured by iohexol (development and validation 2 populations) and 51Cr-EDTA clearances (validation populations 1 and 3) as detailed in the Supplementary data (Supplementary methods and Table S1).
Calculation of eGFR
Since the CKD-EPICr2021 equation has not been validated in non-US populations [6, 15, 27], we additionally considered CKD-EPICr2009 in the analyses [2]. For patients of Caribbean or African origin, we did not apply the CKD-EPICr2009 correction factor for African-American ethnicity, except when specified in subgroup analyses. The latter factor has been shown to overestimate GFR in African Europeans and in sub-Saharan Africans [15, 28, 29]. We also considered GFR estimated by European Kidney Function Consortium (EKFC) [3], CKD-EPICr-Cys2021 [6], CKD-EPICys [30] and FAScombi (Full Age Spectrum equation, based on creatinine and cystatin c) equations [31] (Supplementary data, Table S2)
CT scan protocols
See Supplementary data.
Measurement of skeletal muscle cross-sectional area at the middle of the third lumbar vertebra (L3)
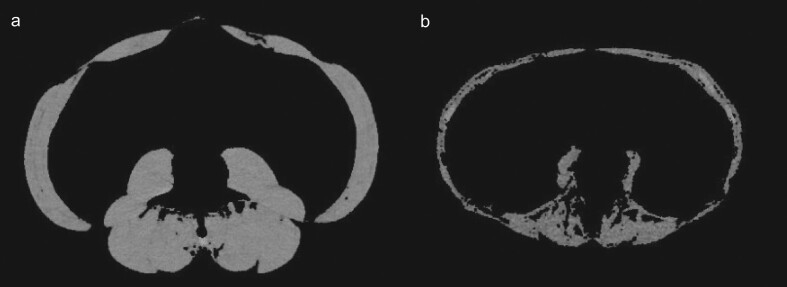
The muscle mass was determined using segmentation of CT scan on dedicated post-treatment station (Advantage Window v4.7; GE Healthcare). A pre-established attenuation threshold of −29 to +150 Hounsfield units was selected and MCSA—defined as the total lumbar MCSA (including the external and internal obliques, paraspinal, rectus abdominis, transversus abdominis and psoas muscles) measured on an axial section passing through the middle of L3—was segmented [16]. Figure 1 illustrates this muscle segmentation in two patients: one with high muscle mass and one with small muscle mass. The interobserver agreement of this measure, assessed in 30 subjects from the development population, was almost perfect: Lin's concordance correlation coefficient = 0.999 (0.998 to 1.0) (Supplementary data, Fig. S1).
Figure 1:
Unenhanced CT scan section taken at the level of the middle of the third lumbar vertebra, after segmentation of total lumbar muscle cross-sectional area. (a) A 61-year-old man with localized kidney cancer. Total lumbar MCSA is 208 cm2. (b) A 70-year-old, kidney donor candidate woman whose past medical history includes only parathyroidectomy for primary hyperparathyroidism complicated by osteoporosis. Total lumbar MCSA is 84 cm2.
Statistical analysis
Continuous variables were expressed as median and interquartile ranges (IQR) or mean and standard deviation (SD), as appropriate. We analyzed, using Pearson test, the correlation of MCSA with urinary creatinine excretion rate, and then with creatinine excreted through glomerular filtration. Because creatinine is freely filtered through the glomerulus, the rate of creatinine excreted after glomerular filtration (U-CreatFiltr) is equal to the filtered load of creatinine [measured GFR (mGFR) × plasma creatinine]. In order to develop a new muscle mass–based eGFR equation, we used multivariable linear regression models to assess the relationship between U-CreatFiltr and the predicting variables, and then we divided the predicted U-CreatFiltr by plasma creatinine (see Supplementary data). We also tested fractional polynomial model for predicting U-CreatFiltr and we compared the goodness-of-fit of linear regression versus fractional polynomial models using Akaike, Bayesian information criterion (BIC) indices and deviance differences. To evaluate the performance of equations, we defined mean bias as the mean difference between eGFR and mGFR, precision as the SD of the bias, and accuracy as the percentage of eGFR values falling within 20% and 30% of mGFR. Visual representations of the agreements were provided as Bland–Altman plots [32]. Some confidence intervals could be wide because of the small size of some validation groups, which would lead to having an upper limit of the 95% confidence interval (95% CI) just above zero, or a lower limit just below, just because of the large width of the 95% CI. Therefore, for eGFR to be categorized as unbiased, not only did the 95% CI have to include zero, but also the bias had to be between −2.5 and +2.5 mL/min/1.73 m2. We compared precisions using Pitman's test for comparison of variances of correlated samples. McNemar's test was used to compare accuracies. Agreement between the methods and mGFR was assessed by Lin's concordance correlation coefficient (CCC) [33]. Statistical analyses were conducted with XLSTAT software (Addinsoft 2021) and Stata v15.0 (StataCorp, College Station, TX, USA).
RESULTS
Clinical characteristics
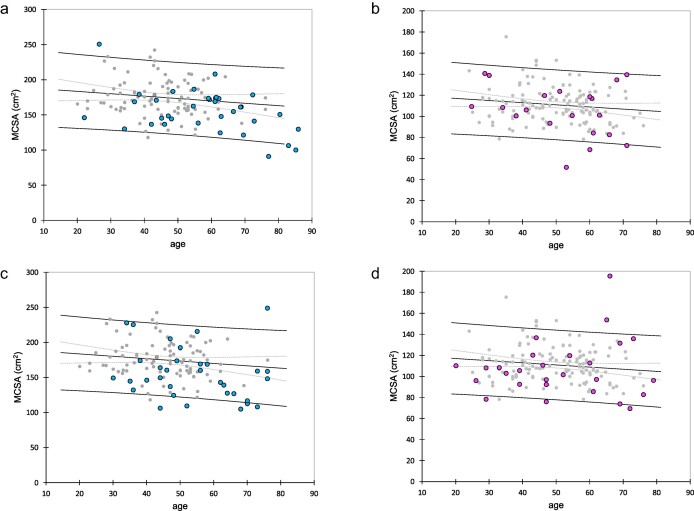
There were 118, 114, 55 and 60 subjects in the development population, and validation populations 1, 2 and 3, respectively. Two patients eligible for validation population 2 were not included because of limb amputation. Clinical characteristics of the patients are detailed in Table 1. The relationship between MCSA and age in the healthy development and validation population 1 subjects is shown as dot plots (gray dots) (Fig. 2). Five patients (9%) and 11 patients (18%) in validation population 2 and 3, respectively, had MCSA outside the 95% prediction interval of the linking linear regression, confirming the high proportion of patients with aberrant muscle mass in validation population 3 (Fig. 2).
Table 1:
Characteristics of participants of the development population and the validation populations.
| Development population | Validation population 1 | Validation population 2 | Validation population 3 | |
|---|---|---|---|---|
| Number of patients | 118 | 114 | 55 | 60 |
| Age, years, mean ± SD | 50.6 ± 12.4 | 45.1 ± 10.6* | 55.5 ± 15.7* | 52.7 ± 15.2 |
| Female, N (%) | 75 (63) | 62 (54) | 20 (36)* | 27 (45)* |
| African or Caribbean ancestry, N (%) | 32 (27) | 14 (12)* | 20 (36) | 14(23) |
| Body weight, kg, mean ± SD | 74.0 ± 14.3 | 73.1 ± 13.6 | 76.7 ± 19.2 | 72.9 ± 18.3 |
| Height, cm, median (IQR) | 168 (160–175) | 168 (161–176) | 170 (163–175) | 170 (160–177) |
| BMI, kg/m2, mean ± SD | 26.1 ± 3.9 | 25.5 ± 4.1 | 26.5 ± 6.0 | 26.0 ± 6.0 |
| Associated diseases, N (%) | ||||
| None | 77 (65) | 87 (76) | 2 (3.6)* | 3 (5)* |
| High blood pressure | 24 (20) | 9 (7.8)* | 33 (60)* | 25 (45)* |
| Obesity (BMI > 30) | 21 (18) | 17 (15) | 14 (25)* | 12(22) |
| Cardiomyopathy | 0 (0) | 0 (0) | 12 (22)* | 8 (13)* |
| Diabetes mellitus | 0 (0) | 0 (0) | 10 (18)* | 15 (25)* |
| Progressive or recent cancer or hematological disease | 0 (0) | 0 (0) | 10 (18)* | 7 (12)* |
| Liver cirrhosis | 0 (0) | 0 (0) | 7 (13)* | 6 (10)* |
| Kidney transplantation | 0 (0) | 0 (0) | 6 (11)* | 18 (30)* |
| Liver transplantation | 0 (0) | 0 (0) | 5 (9.1)* | 4 (6.7)* |
| Neurological disorders | 0 (0) | 0 (0) | 5(9.1)* | 6 (10)* |
| Sickle cell disease | 0 (0) | 0 (0) | 4 (7.2)* | 1 (1.7) |
| Glomerular disease | 0 (0) | 0 (0) | 3 (5.5)* | 3 (5)* |
| Polycystic kidney disease | 0 (0) | 0 (0) | 3 (5.5)* | 2 (3.3)* |
| Malabsorption or digestive resection | 0 (0) | 0 (0) | 3 (5.5)* | 1 (1.7) |
| Inflammatory rheumatism | 0 (0) | 1 (0.9) | 2 (3.6)* | 0(0) |
| HIV seropositive subject | 0 (0) | 0 (0) | 2 (3.6)* | 8 (13)* |
| History of bariatric surgery | 2 (1.7) | 1 (0.8) | 1 (1.8) | 0(0) |
| Lung disease | 0 (0) | 2 (1.8) | 1 (1.8) | 5 (8.3)* |
| Autoimmune disorder | 0 (0) | 0 (0) | 1 (1.8) | 6 (10)* |
| Psychiatric disease | 1 (0.8) | 2 (1.8) | 1 (1.8) | 2 (3.3) |
| mGFR (mL/min/1.73 m2) | 97.4 ± 18.8 | 95.9 ± 13.6 | 60.9 ± 30.5* | 54.6 ± 21.7* |
| Total lumbar MCSA, cm2, median (IQR) | 124 (106–159) | 128 (108–169) | 139 (109–166) | 127 (105–158) |
P-values were calculated between each validation population and development population using a Chi2 test for categorial variables and t-test or Mann–Whitney test as appropriate for quantitative variables. *P < .05.
BMI, body mass index.
Figure 2:
Association between MCSA and age in both sexes. Gray dots represent healthy subjects from the development population and the validation population 1. Linear regression lines in both sexes are shown as well as the 95% prediction limits. Patients from validation population 2 are plotted in (a) (males, blue dots) and (b) (females, pink dots). Patients from validation population 3 are plotted in (c) (males) and (d) (females).
Correlation between urinary creatinine excretion rate and total lumbar MCSA
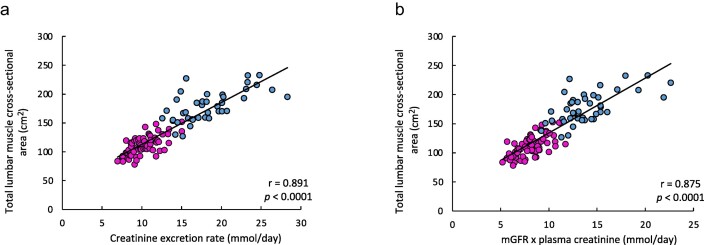
The correlation of urinary creatinine excretion rate with MCSA was strong, r value = 0.891 (P < .0001) (Fig. 3a). The correlation of U-CreatFiltr with MCSA (Fig. 3b) was also very high (r value 0.875, P < .0001).
Figure 3:
Correlation between creatinine urinary excretion and muscle mass assessed by CT scan, in the development population. Correlation between creatinine excretion rate and total lumbar MCSA (a), and between urinary creatinine excretion derived from glomerular filtration (mGFR × plasma creatinine) and MCSA (b). Pink and blue dots represent women and men, respectively. The solid lines represent the linear regressions between the variables. The Pearson correlation coefficient r are reported on the graphs, with the related P-values.
Development of the muscle mass–based eGFR equation (MMB-eGFR)
We considered MCSA as the explanatory variable in the linear regression models where U-CreatFiltr was the dependent variable. Because sex, height and weight displayed collinearity with MCSA, these three variables were not included in the multiple linear regression analyses. The simple linear regression between U-CreatFiltr and MCSA was determined as follows: U-CreatFiltr=– 522.4 + 55.8 × MCSA (R2 = 0.808, RMSE = 980.9). Adding age alone marginally improved the model (R2 = 0.814, RMSE = 971.9). U-CreatFiltr was higher in African and Caribbean descendants [8.2 (7.8–9.9) vs 7.9 (6.7 to 8.7) mmol/day in women, P = .04; and 15.2 (13.9–17.5) vs 12.5 (11.8 to 14.6) mmol/day in men, P = .003]; however, such statistical associations disappeared in the multivariable analysis using MCSA as the primary variable (P = .51). The best fractional polynomial (FP) modeling for U-CreatFiltr indicated a 0.5 power for the best fit but did not increase Akaike indices [Akaike: 1896 for both FP and linear regression (LR)], only slightly improved BIC indices (BIC: 1904 for FP vs 1901 for LR) and did not provide any significant improvement when comparing deviances (P = .997). Therefore, we kept the more parsimonious model, i.e the linear regression model, which retained only MCSA and U-CreatFiltr. MMB-eGFR was written as follows: (−522.4 + 55.8 × MCSA (cm2))/plasma creatinine (µmol/L) and was subsequently adjusted to body surface area [34].
Evaluation of MMB-eGFR equation (vs mGFR) in the development, validation 1, validation 2 and validation 3 populations
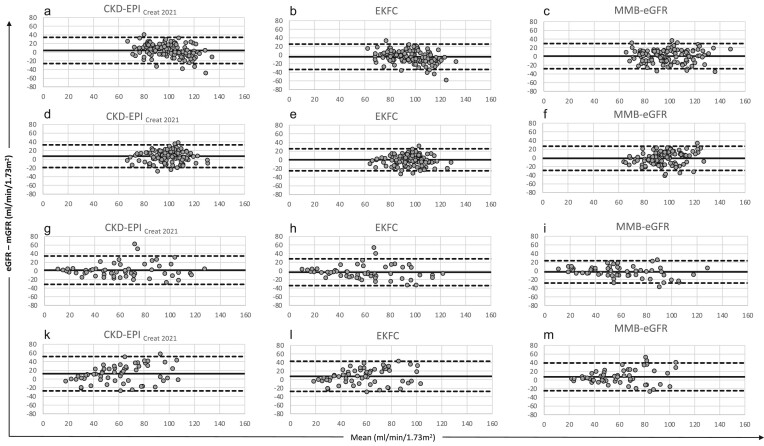
In the development population, both MMB-eGFR and CKD-EPICr2009 equations were unbiased, unlike CKD-EPICr2021 and EKFC, which overestimated and underestimated mGFR, respectively. The precision and accuracy of MMB-eGFR were not different from those of the creatinine and demographic-based equations (Table 2, Fig. 4).
Table 2:
Assessing the performance of MMB-eGFR equation (vs mGFR) in the four study populations, compared with those of equations based on demographic data.
| Mean bias (95% CI) (mL/min/1.73 m2) | SD of the bias (mL/min/1.73 m2) | Accuracy within 20% (95% CI) (%) | Accuracy within 30% (95% CI) (%) | Lin's CCC (95% CI) | |
|---|---|---|---|---|---|
| Development population (n = 118) | |||||
| MMB-eGFR | 0.8 (−1.9 to 3.4) | 14.7 | 85.6 (79.3 to 91.9) | 95.8 (92.1 to 99.4) | 0.69 (0.58 to 0.77) |
| CKD-EPICr2009 | 0.7 (−2.1 to 3.5) | 15.5 | 79.7 (72.4 to 86.9) | 93.2 (88.7 to 97.8) | 0.60 (0.45 to 0.68) |
| CKD-EPICr2021 | 4.0 (1.2 to 6.8) | 15.4 | 76.3 (68.6 to 84.0) | 90.7 (85.4 to 95.9) | 0.59 (0.42 to 0.65) |
| EKFC | −4.0 (−6.6 to −1.2) | 15.0 | 82.2 (75.3 to 89.1) | 94.1 (89.8 to 98.3) | 0.62 (0.46 to 0.68) |
| Validation population 1 (n = 114) | |||||
| MMB-eGFR | −1.1 (−3.7 to 1.5) | 14.2 | 84.2 (77.5 to 90.9) | 96.5 (93.1 to 99.9) | 0.56 (0.42 to 0.67) |
| CKD-EPICr2009 | 3.7 (1.2 to 6.2) | 13.6 | 84.2 (77.5 to 90.9) | 96.5 (93.1 to 99.9) | 0.52 (0.38 to 0.64) |
| CKD-EPICr2021 | 7.0 (4.5 to 9.4) | 13.3 | 73.7 (65.6 to 81.8) | 96.5 (93.1 to 99.9) | 0.48 (0.34 to 0.59) |
| EKFC | 0.1 (−2.3 to 2.5) | 13.0 | 88.6 (82.8 to 94.4) | 98.2 (95.8 to 100) | 0.55 (0.41 to 0.67) |
| Validation population 2 (n = 55) | |||||
| MMB-eGFR | −2.3 (−5.8 to 1.6) | 13.1 | 65.5 (52.9 to 78.0) | 80.0 (69.4 to 90.6) | 0.89 (0.83 to 0.93) |
| CKD-EPICr2009 | −1.1 (−5.4 to 3.3) | 16.5* | 56.4 (43.3 to 69.5) | 80.0 (69.4 to 90.6) | 0.85 (0.76 to 0.91) |
| CKD-EPICr2021 | 1.6 (−2.7 to 6.1) | 16.8* | 61.8 (49.0 to 74.7) | 80.0 (69.4 to 90.6) | 0.85 (0.76 to 0.91) |
| EKFC | −3.1 (−7.3 to 1.1) | 15.9* | 56.4 (43.3 to 69.5) | 83.6 (73.9 to 94.4) | 0.85 (0.76 to 0.91) |
| Validation population 3 (n = 60) | |||||
| MMB-eGFR | 7.4 (3.2 to 11.5) | 16.3 | 56.7 (44.1 to 69.2) | 75.0 (64.0 to 86.0) | 0.72 (0.58 to 0.82) |
| CKD-EPICr2009 | 9.5 (4.4 to 14.5) | 20.0* | 30.0 (18.4 to 41.6)* | 51.7 (39.0 to 64.3)* | 0.63 (0.46 to 0.75) |
| CKD-EPICr2021 | 12.3 (7.2 to 17.4) | 20.2* | 30.0 (18.4 to 41.6)* | 43.3 (31.0 to 55.9)* | 0.60 (0.44 to 0.72) |
| EKFC | 7.7 (3.1 to 12.2) | 18.0 | 35.0 (24.5 to 48.9)* | 53.3 (40.7 to 66.0)* | 0.67 (0.52 to 0.78) |
P-values were calculated between each eGFR equation based on age and sex, and MMB-eGFR, in each population. *P < .05. The precision (SD of the bias) comparison was performed with Pitman's test. Accuracy comparison was performed with McNemar's test.
Lin's CCC, Lin's concordance correlation coefficient.
Figure 4:
Bland and Altman comparison between creatinine based eGFR and mGFR, in the four studied populations: (a–c) development population; (d–f) validation population 1; (g–i) validation population 2; (k–m) validation population 3. CKD-EPICreat2021, EKFC and MMB-eGFR, respectively. The solid line indicates the mean bias, and dashed lines indicate the lower and upper limits of the agreement interval (−1.96 SD and +1.96 SD). All results are in mL/min/1.73 m2.
In validation population 1, CKD-EPICr2009 and CKD-EPICr2021 were positively biased, whereas MMB-eGFR and EKFC were unbiased (Table 2, Fig. 4). Precision and accuracy of MMB-eGFR were not different from those of other eGFR equations (Table 2, Fig. 4). In validation population 2, the biases of MMB-eGFR, CKD-EPICr2009 and CKD-EPICr2021 were not significantly different from zero, whereas that of EKFC was negative (Table 2, Fig. 4), and MMB-eGFR showed better precision than all other equations (Table 2, Fig. 4). In validation population 3, all equations had positive biases. Precision and accuracy of MMB-eGFR were superior to those of CKD-EPICr2009, CKD-EPICr2021 and EKFC (Table 2, Fig. 4).
Comparing MMB-eGFR with equations based on cystatin C
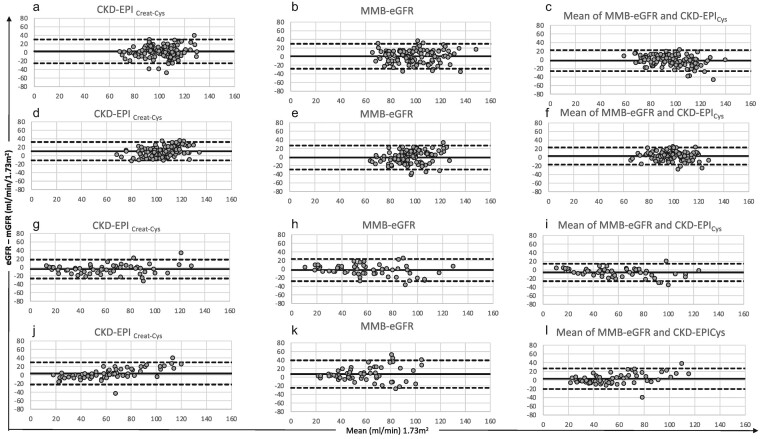
We compared the performance of MMB-eGFR with that of CKD-EPICys, CKD-EPICr-Cys2021 and FAScombi, and evaluated the performance of the mean of MMB-eGFR and CKD-EPICys equations, an approach combining the two endogenous GFR markers and incorporating muscle mass (Table 3, Fig. 5) (the limited size of the development population, which did not include any patients with low GFR values, did not allow us to develop an equation incorporating both creatinine and cystatin C). CKD-EPICys showed opposite biases between the two groups of healthy subjects: development population (Roche assay, negative bias) and validation population 1 (Siemens assay, positive bias). CKD-EPICr-Cys2021 and FAScombi were positively and negatively biased in validation populations 1 and 2, respectively. CKD-EPICr-Cys2021 had higher precision than that of MMB-eGFR in validation population 1. The precision of the mean of MMB-eGFR and CKD-EPICys was higher than that of CKD-EPICr-Cys2021 in development population, and higher than that of FAScombi in all populations except in validation population 3. The accuracy of the mean of MMB-eGFR and CKD-EPICys was higher than that of CKD-EPICr-Cys2021 and FAScombi in validation population 1 (higher accuracy within 30% for both equations, higher accuracy within 20% for CKD-EPICr-Cys2021).
Table 3:
Assessing the performance of MMB-eGFR and of the mean of MMB-eGFR and CKD-EPICys equations (vs mGFR) in the four study populations, compared with those of the CKD-EPICys, CKD-EPICr-Cys2021 and FAScombi equations.
| Mean bias (95% CI) (mL/min/1.73 m2) | SD of the bias (mL/min/1.73 m2) | Accuracy within 20% (95% CI) (%) | Accuracy within 30% (95% CI) (%) | Lin's CCC (95% CI) | |
|---|---|---|---|---|---|
| Development population (n = 117) | |||||
| MMB-eGFR | 0.8 (−1.9 to 3.5) | 14.8 | 85.5 (79.1 to 91.9) | 95.7 (92.1 to 99.4) | 0.69 (0.58 to 0.77) |
| Mean MMB-eGFR/CKD-EPICys | −2.0 (−4.2 to 0.2) | 12.5 | 88.9 (83.2 to 94.6) | 99.1 (97.5 to 100) | 0.73 (0.64 to 0.80) |
| CKD-EPICys | −5.0 (−8.0 to −2.0) | 16.6 | 78.6 (71.2 to 86.1) | 91.5 (86.4 to 96.5) | 0.56 (0.43 to 0.67) |
| CKD-EPICr-Cys2021 | 2.5 (0.0 to 5.1) | 14.2† | 80.3 (73.1 to 87.5) | 94.9 (90.9 to 98.9) | 0.64 (0.53 to 0.73) |
| FAScombi | −4.5 (−7.2 to −1.7) | 15.0† | 83.8 (77.1 to 90.4) | 94.0 (89.7 to 98.3)† | 0.58 (0.45 to 0.69) |
| Validation population 1 (n = 110) | |||||
| MMB-eGFR | −1.1 (−3.7 to 1.5) | 14.4 | 83.6 (76.7 to 90.6) | 96.4 (92.9 to 99.9) | 0.56 (0.42 to 0.67) |
| Mean MMB-eGFR/CKD-EPICys | 3.0 (1.1 to 4.9) | 10.2 | 90.0 (84.4 to 99.6) | 99.1 (97.3 to 100) | 0.69 (0.58 to 0.77) |
| CKD-EPICys | 7.1 (4.7 to 9.4) | 12.7 | 76.4 (68.4 to 84.3) | 92.7 (87.9 to 97.6) | 0.54 (0.41 to 0.64) |
| CKD-EPICr-Cys2021 | 10.6 (8.5 to 12.6) | 11.1* | 72.7 (64.4 to 81.1)† | 90.9 (85.5 to 96.3)† | 0.51 (0.39 to 0.61) |
| FAScombi | 4.5 (1.8 to 7.2) | 14.5† | 81.8 (74.6 to 89.0) | 92.7 (87.9 to 97.6)† | 0.57 (0.43 to 0.67) |
| Validation population 2 (n = 54) | |||||
| MMB-eGFR | −2.3 (−5.8 to 1.2) | 13.2 | 64.8 (52.1 to 77.6) | 79.6 (68.9 to 90.4) | 0.89 (0.83 to 0.93) |
| Mean MMB-eGFR/CKD-EPICys | −5.9 (−8.7 to −3.1) | 10.4 | 64.8 (52.1 to 77.6) | 94.4 (88.3 to 100) | 0.92 (0.87 to 0.95) |
| CKD-EPICys | −9.5 (−13.0 to −6.0) | 13.2 | 46.3 (33.0 to 59.6) | 75.9 (64.5 to 87.3) | 0.86 (0.78 to 0.92) |
| CKD-EPICr-Cys2021 | −4.1 (−7.1 to −1.1) | 11.4 | 66.7 (54.1 to 79.2) | 87.0 (78.1 to 96.0) | 0.92 (0.87 to 0.95) |
| FAScombi | −5.2 (−8.6 to −1.8) | 12.7† | 63.0 (50.1 to 75.8) | 83.3 (73.4 to 93.3)† | 0.88 (0.80 to 0.93) |
| Validation population 3 (n = 60) | |||||
| MMB-eGFR | 7.4 (3.2 to 11.5) | 16.3 | 56.7 (44.1 to 69.2) | 75.0 (64.0 to 86.0) | 0.72 (0.58 to 0.82) |
| Mean MMB-eGFR/CKD-EPICys | 3.2 (0.1 to 6.2) | 12.1 | 71.7 (60.3 to 83.1) | 86.7 (78.1 to 95.3) | 0.87 (0.81 to 0.92) |
| CKD-EPICys | −1.0 (−6.2 to 4.2) | 20.5 | 40.0 (27.6 to 52.4) | 61.7 (49.4 to 74.0) | 0.75 (0.65 to 0.82) |
| CKD-EPICr-Cys2021 | 3.8 (0.4 to 7.1) | 13.3 | 56.7 (44.1 to 69.2)† | 81.7 (71.9 to 91.5) | 0.85 (0.78 to 0.90) |
| FAScombi | 3.3 (0.5 to 6.2) | 11.3* | 66.7 (54.7 to 78.6) | 78.3 (67.9 to 88.8) | 0.87 (0.79 to 0.92) |
P-values were calculated between each eGFR equation based on age and sex (CKD-EPICys, CKD-EPICr-Cys2021 and FAScombi) and MMB-eGFR, in each population. *P < .05. For CKD-EPICr-Cys2021 and FAScombi, P-values were also calculated against Mean MMB-eGFR/CKD-EPICys in each population. †P < .05. The precision (SD of the bias) comparison was performed with Pitman's test. Accuracy comparison was performed with McNemar's test.
Lin's CCC, Lin's concordance correlation coefficient.
Figure 5:
Bland and Altman analysis between creatinine and cystatin based eGFR and mGFR, in the four populations: (a–c) development population; (d–f) validation population 1; (g–i) validation population 2; (j–l) validation population 3. CKD-EPICreat-Cys, MMB-eGFR, mean of MMB-eGFR and CKD-EPICys, respectively. The solid line indicates the mean bias, and the dashed lines indicate the lower and upper limits of the agreement interval (−1.96 SD and +1.96 SD). All results are in mL/min/1.73 m2.
Evaluation of MMB-eGFR equation (vs mGFR) in patients stratified by ethnicity
Because hypothetical differences in body composition due to ethnicity could be attenuated or exacerbated by aberrant muscle masses related to the selection criteria of validation population 3, we did not include these patients in this analysis. Thirty-four patients of validation populations 1 and 2 were of African or Caribbean origin, and 135 were non-Black. Although MMB-eGFR slightly underestimated mGFR in subjects of African or Caribbean origin, the difference in bias between Black and white subjects was very small and not statistically different from zero: −2.1 mL/min/1.73 m2 (95% CI −7.2 to 3.0). The difference in biases were greater for CKD-EPICr-2009, CKD-EPICr-2021, EKFC and CKD-EPICr-Cys2021 [−8.2 mL/min/1.73 m2 (−13.0 to −3.4); −8.4 mL/min/1.73 m2 (−13.2 to −3.6); −6.3 mL/min/1.73 m2 (−10.9 to −1.8); and −3.8 mL/min/1.73 m2 (−8.6 to 1.0)] (Table 4). We also performed additional analyses to determine whether any differences in muscle mass by ethnicity influenced serum creatinine levels. For this purpose, we selected only healthy subjects (development population and validation population 1). We matched each Black subject (n = 41) with two randomly selected white subjects of the same age and sex (n = 82). The ratio of median plasma creatinine between Black and non-Black subjects was greater in men than in women (124% and 108%) (Supplementary data, Table S3). The ratios of urinary creatinine excretion were of the same magnitude as those of plasma creatinine, which suggests that the differences in plasma creatinine were attributable to differences in creatinine generation. Also supporting this hypothesis is the absence of differences in tubular creatinine secretion between ethnic groups (Supplementary data, Table S3). MCSA accounted almost entirely for this difference in urinary creatinine excretion (Supplementary data, Table S3 and Fig. S2). Besides differences in muscle mass, there could also be differences in creatinine metabolism, or in diet. These hypotheses were not investigable, because patients should have been matched not only for age and sex but also for weight, height (and/or MCSA) and mGFR, which was not possible in this study.
Table 4:
Assessing the performance of MMB-eGFR equation (vs mGFR) in patients of African or Caribbean origin and in non-Black patients, compared with those of equations based on demographic data.
| Mean bias (95% CI) (mL/min/1.73 m2) | SD of the bias (mL/min/1.73 m2) | Accuracy within 20% (95% CI) (%) | Accuracy within 30% (95% CI) (%) | Lin's CCC (95% CI) | |
|---|---|---|---|---|---|
| Patients with African or Caribbean ancestry (n = 34: V1 = 14; V2 = 20) | |||||
| MMB-eGFR | −3.2 (−7.7 to 1.4) | 13.5 | 79.4 (65.8 to 93.0) | 91.2 (81.6.to 100) | 0.88 (0.78 to 0.94) |
| CKD-EPICrCr2009 no-race | −4.4 (−8.5 to −0.3) | 12.3 | 70.6 (55.3 to 85.9) | 91.2 (81.6.to 100) | 0.90 (0.82 to 0.95) |
| CKD-EPICrCr2009 race | 6.6 (1.6 to 11.6) | 14.8 | 76.5 (62.2 to 90.7) | 79.4 (65.8 to 93.0) | 0.87 (0.78 to 0.93) |
| CKD-EPICr2021 | −1.4 (−5.5 to 2.6) | 12.2 | 73.5 (58.7 to 88.4) | 94.1 (91.1 to 100) | 0.92 (0.84 to 0.96) |
| EKFC | −6.0 (−9.8 to −2.2) | 11.4 | 73.5 (58.7 to 88.4) | 91.2 (81.6 to 100) | 0.90 (0.81 to 0.95) |
| EKFC with African-European Q values | 0.6 (−3.3 to 4.4) | 11.4 | 76.5 (62.2 to 90.7) | 91.2 (81.6 to 100) | 0.92 (0.85 to 0.96) |
| CKD-EPICys (n = 33) | 2.4 (−2.4 to 7.2) | 14.1 | 60.6 (43.9 to 77.3) | 90.9 (81.1 to 100) | 0.90 (0.82 to 0.94) |
| Mean MMB-eGFR/CKD-EPICys (n = 33) | −0.3 (−3.9 to 3.2) | 10.6 | 81.8 (68.7 to 95.0) | 100 | 0.94 (0.88 to 0.97) |
| CKD-EPICr-Cys2021 (n = 33) | 2.7 (−1.5 to 7.0) | 12.5 | 75.8 (58.4 to 93.2) | 93.9 (85.8 to 100) | 0.92 (0.85 to 0.95) |
| FAScombi (n = 33) | −1.4 (−5.5 to 2.8) | 12.1 | 72.7 (57.5 to 87.9) | 93.9 (85.8 to 100) | 0.91 (0.84 to 0.95) |
| Non-Black patients (n = 135: V1 = 100; V2 = 35) | |||||
| MMB-eGFR | −1.0 (−3.4 to 1.3) | 14.0 | 77.8 (70.8 to 84.8) | 91.1 (86.3 to 95.9) | 0.85 (0.80 to 0.89) |
| CKD-EPICr2009 | 3.8 (1.3 to 6.3) | 14.9 | 76.3 (69.1 to 83.5) | 91.1 (86.3 to 95.9) | 0.83 (0.76 to 0.87) |
| CKD-EPICr2021 | 6.9 (4.4 to 9.4) | 14.8 | 68.9 (61.1 to 76.7) | 89.6 (84.5 to 94.8) | 0.81 (0.74 to 0.86) |
| EKFC | 0.3 (−2.1 to 2.7) | 14.3 | 79.3 (72.4 to 86.1) | 94.1 (90.1 to 98.1) | 0.84 (0.78 to 0.88) |
| CKD-EPICys (n = 131) | 1.4 (−1.2 to 4.0) | 15.3 | 67.9 (60.0 to 75.9) | 86.3 (80.4 to 92.2) | 0.85 (0.80 to 0.89) |
| Mean MMB-eGFR/CKD-EPICys (n = 131) | 0.2 (−1.8 to 2.1) | 11.2 | 81.7 (75.1 to 88.3) | 97.0 (94.0 to 99.9) | 0.91 (0.87 to 0.93) |
| CKD-EPICr-Cys2021 (n = 131) | 6.5 (4.2 to 8.7) | 13.2 † | 69.5 (61.6 to 77.4)† | 88.6 (83.1 to 94.0)† | 0.86 (0.81 to 0.89) |
| FAScombi (n = 131) | 2.0 (−0.6 to 4.6) | 15.2 † | 76.3 (69.1 to 83.6) | 88.6 (83.1 to 94.0)† | 0.84 (0.79 to 0.88) |
For CKD-EPICr2009, CKD-EPICr2021, EKFC and CKD-EPICys, P-values were calculated against MMB-eGFR in each population. *P < .05. For CKD-EPICr-Cys2021 and FAScombi, P-values were calculated against Mean MMB-eGFR/CKD-EPICys, in each population. †P < .05. The precision (SD of the bias) comparison was performed with Pitman's test. Accuracy comparison was performed with McNemar's test.
CKD-EPICr2009 no-race, CKD-EPICr2009 without African American correction factor; CKD-EPICr2009 race, CKD-EPICr2009 with African American correction factor; Lin's CCC, Lin's concordance correlation coefficient; V1/V2, validation population 1/2.
DISCUSSION
We developed and externally validated a new GFR estimation equation that derives its main variables from creatinine plasma level and total lumbar MCSA measured by CT scan. The latter is a surrogate marker of total skeletal muscle mass. MMB-eGFR was as accurate as creatinine- and demographics-based equations in healthy subjects, and had better performances in patients with chronic illnesses, especially if they have atypical muscle masses. This new equation could also be applied to subjects of various ethnic backgrounds, because its bias was only marginally affected by ethnicity.
Using a CT scan–derived surrogate of muscle mass in a creatinine-based GFR estimation equation should be justified beforehand by evidencing that such CT scan data are associated with creatinine generation. We fulfilled this requirement in the first part of the study by showing the good correlation between MCSA and urinary creatinine excretion. Our results confirm those of another recent publication showing that urinary creatinine excretion could be estimated from a combination of various body composition parameters, including psoas, long spine and abdominal wall muscle areas [35]. Bioelectrical impedance (BIA) had also been considered in the past as a tool to improve the accuracy of creatinine-based GFR estimation [36]. While MCSA measurement at the third lumbar vertebra provides an estimate of skeletal muscle mass, BIA estimates lean body mass from whole-body electrical conductivity, using estimating equations based on demographic and/or morphometric data (height, weight, ethnicity) that may differ between instrument brands, the technology used [single vs multi-frequency and multi-segmental instruments using two vs more electrodes (up to eight)], and reference populations used. Hydration status can also influence BIA measurements [37]. Lean body mass assessment by BIA avoid exposure to radiation, but may be limited by the availability of the device and the time required to perform the measurements. To date, total lumbar MCSA measured by CT scan is probably the easiest and most promising tool available in routine care to estimate total skeletal muscle mass, reliable and with excellent inter- and intra-observer agreement [20–23, 38]. One could argue that the cost and radiation of CT scan might discourage its employment for GFR estimation; nevertheless, for the many patients routinely followed up by CT scans, using the MMB-eGFR equation could help improving kidney function assessment without additional procedure, cost or radiation, consistent with the concept of value-added opportunistic CT screening. This concept has been extensively detailed in a recent review [39]: value-added opportunistic CT screening is the practice of using incidental imaging data unrelated to the clinical indication for prevention, risk profiling or presymptomatic detection of relevant disease. When the data are available, we propose to use it, since MMB-eGFR performs as well as (in healthy subjects) or better than (in case of atypical muscle mass) equations based on demographic data. Future research may allow determination of MCSA thresholds below or above which MMB-eGFR should be preferred to demographic-based equations. However, MMB-eGFR is not a GFR measurement. Therefore, when the exact GFR value is required to make a clinical decision, MMB-eGFR should not be used as a substitute for GFR measurement.
Demographic-based equations model creatinine generation in addition to GFR. This feature is highly apparent in the FAS and EKFC equations, which incorporate the median creatinine values for each gender in healthy subjects. In case of aberrant muscle mass, MMB-eGFR, which models personalized creatinine generation based on muscle mass, performs better. Other populations could potentially benefit from this equation such as people transitioning from one sex to the other, and those whose body habitus does not correspond to gender conformity norms. Similarly, if we consider that ethnicity corrective factor for eGFR is meant to overcome the differences in muscle creatinine production between ethnic groups [12, 13], MMB-eGFR could be accurately applied to patients of any ethnic origin. Of note, in the development population, despite higher U-CreatFiltr in Black subjects, ethnicity was not statistically associated with U-CreatFiltr in the multivariable analysis. Moreover, the differences in bias for MMB-eGFR between Black and non-Black patients in the validation populations were small. Finally, in healthy subjects, the differences in MCSA between Black and white subjects accounted almost entirely for differences in both urinary creatinine excretion and plasma creatinine levels. Our approach is in alignment with the recent recommendations of the NKF-ASN task force [5]. The issue of race-specific factors is not limited to African-American subjects: to date, at least 10 different race coefficients have been proposed in creatinine-based GFR estimation equations [40]. Our results add to other efforts to eliminate the use of race coefficients in GFR estimation, such as the development of cystatin C–based GFR estimation equations, which do not require race or sex factors and have been available since 2006 [40–42]. Recent American guidelines also called for a wider use of CKD-EPICr-Cys2021 equation since it has a greater accuracy than CKD-EPICr2021, and a smaller impact of race-based imprecision factors [5]. However, the use of cystatin C–based eGFR is limited by the cost of cystatin C assay, and biases between the measurements of different manufacturers: we also found such differences in bias with CKD-EPICys in the validation population 1 (Siemens test) and in the development population (Roche test) [43, 44]. This imperfect agreement between manufacturers’ assays is noted despite the availability of a reference material, ERM-DA471/IFCC for the cystatin C [45], and is a result of the failure of reagent producers to adopt recommended standardization procedures. However, manufacturers are gradually improving their accuracy [46]. Another limitation of equations based on both creatinine and cystatin C is the impact, albeit small, of race on their performance. That said, MMB-eGFR did not perform better than CKD-EPICr-Cys2021 or FAScombi, but the mean of MMB-eGFR and CKD-EPICys had better precision than FAScombi in validation population 2, and better accucary within 20% than CKD-EPICr-Cys2021 in the validation population 3.
Our study has some limitations. In patients with very low GFR, tubular creatinine secretion is expected to be increased, protein intakes are often decreased (intentionally or not) and extra-renal creatinine clearance may become significant [47, 48]. Having no patient with low GFR in the development population, and very few in the validation groups can therefore be considered as a limitation of our study. Another limitation is the small size of the three validation populations: while MMB-eGFR is probably not superior to the standard creatinine-based equations in healthy subjects, the lack of difference in accuracy in subjects with various chronic illnesses (validation population 2), despite the evidence of better precision of MMB-eGFR, is likely due to insufficient statistical power. Yet, we highlighted a strong benefit of our approach in patients who were at risk of having aberrant muscle masses (validation population 3), demonstrating, in this pilot study, the potential usefulness of using a surrogate marker for total muscle mass in creatinine-based eGFR. Because the selection criteria of validation populations were very different, we did not merge them in order to increase statistical power, except for the analyses adjusted for ethnicity. Finally, the question of the time limit within which CT scan data are valid is not answered. In subjects with no new disease and with stable physical activity and diet, the time limit is probably longer than the 6 months we arbitrarily defined. Inversely, patients developing severe acute diseases could suffer rapid muscle mass reduction [49], which may make their old CT scans unsuitable to estimate their GFR.
In conclusion, the new MMB-eGFR equation estimates GFR using, as the sole supplemental marker in addition to plasma creatinine, muscle mass assessed by total lumbar MCSA (no demographic data). This equation is more accurate than creatinine and demographic-based equations in patients with atypical muscle mass. Furthermore, we showed that MMB-eGFR performance, including its bias, is only marginally affected by ethnicity.
Such results highlighting the relevance of considering muscle mass assessed by CT scan to estimate creatinine-based GFR, in a value-added CT opportunistic screening approach [39], need to be confirmed and refined by large-scale research projects.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the patients, nurses and physicians who participated in the GFR measurement procedures.
Contributor Information
Thomas Stehlé, Univ. Paris Est Créteil, Institut National de la Santé et de la Recherche Médicale (INSERM) U955, Institut Mondor de Recherche Biomédicale (IMRB), Créteil, France; Assistance Publique des Hôpitaux de Paris (AP-HP), Hôpitaux Universitaires Henri Mondor, Service de Néphrologie et Transplantation, Fédération Hospitalo-Universitaire « Innovative therapy for immune disorders », Créteil, France.
Yaniss Ouamri, Univ. Paris Est Créteil, Institut National de la Santé et de la Recherche Médicale (INSERM) U955, Institut Mondor de Recherche Biomédicale (IMRB), Créteil, France; Assistance Publique des Hôpitaux de Paris (AP-HP), Hôpitaux Universitaires Henri-Mondor, Service d'Imagerie Médicale, Créteil, France.
Antoine Morel, Univ. Paris Est Créteil, Institut National de la Santé et de la Recherche Médicale (INSERM) U955, Institut Mondor de Recherche Biomédicale (IMRB), Créteil, France; Assistance Publique des Hôpitaux de Paris (AP-HP), Hôpitaux Universitaires Henri-Mondor, Service de Santé Publique, Créteil, France.
Emmanuelle Vidal-Petiot, Université Paris Cité, Institut National de la Santé et de la Recherche Médicale (INSERM), U1149, Paris, France; Assistance Publique des Hôpitaux de Paris (AP-HP), Département de Physiologie-Explorations Fonctionnelles, Hôpital Bichat, Paris, France.
Soraya Fellahi, Université Pierre et Marie Curie Paris 6, Hôpital Pitié-Salpêtrière, Assistance Publique des Hôpitaux de Paris (APHP), Paris, France; Assistance Publique des Hôpitaux de Paris (AP-HP), Hôpitaux Universitaires Henri Mondor, Département de Biochimie, Créteil, France.
Lauriane Segaux, Univ. Paris Est Créteil, Institut National de la Santé et de la Recherche Médicale (INSERM) U955, Institut Mondor de Recherche Biomédicale (IMRB), Créteil, France; Assistance Publique des Hôpitaux de Paris (AP-HP), Hôpitaux Universitaires Henri-Mondor, Service de Santé Publique, Créteil, France.
Dominique Prié, Université de Paris Cité, Faculté de Médecine, Institut National de la Santé et de la Recherche Médicale (INSERM) U1151, Paris, France; Assistance Publique des Hôpitaux de Paris (AP-HP), Groupe Hospitalier Necker Enfants Malades, Service de Physiologie et Explorations Fonctionnelles, Paris, France.
Philippe Grimbert, Univ. Paris Est Créteil, Institut National de la Santé et de la Recherche Médicale (INSERM) U955, Institut Mondor de Recherche Biomédicale (IMRB), Créteil, France; Assistance Publique des Hôpitaux de Paris (AP-HP), Hôpitaux Universitaires Henri Mondor, Service de Néphrologie et Transplantation, Fédération Hospitalo-Universitaire « Innovative therapy for immune disorders », Créteil, France.
Alain Luciani, Univ. Paris Est Créteil, Institut National de la Santé et de la Recherche Médicale (INSERM) U955, Institut Mondor de Recherche Biomédicale (IMRB), Créteil, France; Assistance Publique des Hôpitaux de Paris (AP-HP), Hôpitaux Universitaires Henri-Mondor, Service d'Imagerie Médicale, Créteil, France.
Vincent Audard, Univ. Paris Est Créteil, Institut National de la Santé et de la Recherche Médicale (INSERM) U955, Institut Mondor de Recherche Biomédicale (IMRB), Créteil, France; Assistance Publique des Hôpitaux de Paris (AP-HP), Hôpitaux Universitaires Henri Mondor, Service de Néphrologie et Transplantation, Fédération Hospitalo-Universitaire « Innovative therapy for immune disorders », Créteil, France.
Jean Philippe Haymann, Univ. Paris Diderot, Sorbonne Paris Cité, Institut National de la Santé et de la Recherche Médicale (INSERM), U1155; Assistance Publique des Hôpitaux de Paris (AP-HP), Hôpitaux de Paris, hôpital Tenon, Département de Physiologie-Explorations Fonctionnelles, Paris, France.
Sébastien Mulé, Univ. Paris Est Créteil, Institut National de la Santé et de la Recherche Médicale (INSERM) U955, Institut Mondor de Recherche Biomédicale (IMRB), Créteil, France; Assistance Publique des Hôpitaux de Paris (AP-HP), Hôpitaux Universitaires Henri-Mondor, Service d'Imagerie Médicale, Créteil, France.
Eric De Kerviler, Assistance Publique des Hôpitaux de Paris (AP-HP), Hôpitaux de Paris, Hôpital Tenon, Département de Physiologie-Explorations Fonctionnelles, Paris, France.
Marie-Noëlle Peraldi, Assistance Publique des Hôpitaux de Paris (AP-HP), Hôpital Saint Louis, Service de Néphrologie, Paris, France.
Anne Boutten, Assistance Publique des Hôpitaux de Paris (AP-HP), Hôpitaux de Paris, hôpital Bichat, Département de Biochimie Clinique, Paris, France.
Marie Matignon, Univ. Paris Est Créteil, Institut National de la Santé et de la Recherche Médicale (INSERM) U955, Institut Mondor de Recherche Biomédicale (IMRB), Créteil, France; Assistance Publique des Hôpitaux de Paris (AP-HP), Hôpitaux Universitaires Henri Mondor, Service de Néphrologie et Transplantation, Fédération Hospitalo-Universitaire « Innovative therapy for immune disorders », Créteil, France.
Florence Canouï-Poitrine, Univ. Paris Est Créteil, Institut National de la Santé et de la Recherche Médicale (INSERM) U955, Institut Mondor de Recherche Biomédicale (IMRB), Créteil, France; Assistance Publique des Hôpitaux de Paris (AP-HP), Hôpitaux Universitaires Henri-Mondor, Service de Santé Publique, Créteil, France.
Martin Flamant, Université Paris Cité, Institut National de la Santé et de la Recherche Médicale (INSERM), U1149, Paris, France; Assistance Publique des Hôpitaux de Paris (AP-HP), Département de Physiologie-Explorations Fonctionnelles, Hôpital Bichat, Paris, France.
Frédéric Pigneur, Univ. Paris Est Créteil, Institut National de la Santé et de la Recherche Médicale (INSERM) U955, Institut Mondor de Recherche Biomédicale (IMRB), Créteil, France; Assistance Publique des Hôpitaux de Paris (AP-HP), Hôpitaux Universitaires Henri-Mondor, Service d'Imagerie Médicale, Créteil, France.
AUTHORS’ CONTRIBUTIONS
T.S. and F.P. conceived and designed the study and drafted the first version of the manuscript. Y.O. performed CT scan segmentations to measure muscle mass and participated in participated in the interpretation of the data. T.S., A.M., L.S. and F.C.-P. performed statistical analyses. E.V.-P., S.F., D.P., A.L., S.M., E.D.K. and M.F. contributed to the acquisition of clinical biological or radiological data and critically reviewed the manuscript. P.G., V.A., J.P.H., S.M. and M.N.P. participated in the interpretation of the data and critically reviewed the manuscript. All authors contributed important intellectual content during manuscript drafting and approved the submitted version of the manuscript
DATA AVAILIBILITY STATEMENT
The data underlying this article will be shared on reasonable request to the corresponding author.
CONFLICT OF INTEREST STATEMENT
All the authors declare no competing interests.
REFERENCES
- 1. Levey AS, Bosch JP, Lewis JBet al. . A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of diet in Renal Disease Study Group. Ann Intern Med 1999;130:461–70. [DOI] [PubMed] [Google Scholar]
- 2. Levey AS, Stevens LA, Schmid CHet al. . A new equation to estimate glomerular filtration rate. Ann Intern Med 2009;150:604–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pottel H, Björk J, Courbebaisse Met al. . Development and validation of a modified full age spectrum creatinine-based equation to estimate glomerular filtration rate: a cross-sectional analysis of pooled data. Ann Intern Med 2021;174:183–91. [DOI] [PubMed] [Google Scholar]
- 4. Ku E, McCulloch CE, Adey DBet al. . Racial disparities in eligibility for preemptive waitlisting for kidney transplantation and modification of eGFR thresholds to equalize waitlist time. J Am Soc Nephrol 2021;32:677–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Delgado C, Baweja M, Burrows NRet al. . Reassessing the inclusion of race in diagnosing kidney diseases: an interim report from the NKF-ASN Task Force. J Am Soc Nephrol 2021;32:1305–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Inker LA, Eneanya ND, Coresh Jet al. . New creatinine- and cystatin C–based equations to estimate GFR without race. N Engl J Med 2021;385:1737–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Myers GL. Recommendations for improving serum creatinine measurement: a report from the Laboratory Working Group of the National Kidney Disease Education Program. Clin Chem 2006;52:5–18. [DOI] [PubMed] [Google Scholar]
- 8. Stevens LA, Levey AS.. Measured GFR as a confirmatory test for estimated GFR. J Am Soc Nephrol 2009;20:2305–13. [DOI] [PubMed] [Google Scholar]
- 9. Jacobsen FK, Christensen CK, Mogensen CEet al. . Pronounced increase in serum creatinine concentration after eating cooked meat. BMJ 1979;1:1049–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shah KF, Stevens PE, Lamb EJ.. The influence of a cooked-fish meal on estimated glomerular filtration rate. Ann Clin Biochem 2020;57:182–5. [DOI] [PubMed] [Google Scholar]
- 11. Garimella PS, Tighiouart H, Sarnak MJet al. . Tubular secretion of creatinine and risk of kidney failure: the Modification of Diet in Renal Disease (MDRD) study. Am J Kidney Dis 2021;77:992–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ix JH, Wassel CL, Stevens LAet al. . Equations to estimate creatinine excretion rate: the CKD Epidemiology Collaboration. Clin J Am Soc Nephrol 2011;6:184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ortiz O, Russell M, Daley TLet al. . Differences in skeletal muscle and bone mineral mass between black and white females and their relevance to estimates of body composition. Am J Clin Nutr 1992;55:8–13. [DOI] [PubMed] [Google Scholar]
- 14. Hsu C, Yang W, Parikh RVet al. . Race, genetic ancestry, and estimating kidney function in CKD. N Engl J Med 2021;385:1750–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delanaye P, Vidal-Petiot E, Björk Jet al. . Performance of creatinine-based equations to estimate glomerular filtration rate in White and Black populations in Europe, Brazil and Africa. Nephrol Dial Transplant 2023;38:106–18. [DOI] [PubMed] [Google Scholar]
- 16. Mourtzakis M, Prado CMM, Lieffers JRet al. . A practical and precise approach to quantification of body composition in cancer patients using computed tomography images acquired during routine care. Appl Physiol Nutr Metab 2008;33:997–1006. [DOI] [PubMed] [Google Scholar]
- 17. Shen W, Punyanitya M, Wang Zet al. . Total body skeletal muscle and adipose tissue volumes: estimation from a single abdominal cross-sectional image. J Appl Physiol 2004;97:2333–8. [DOI] [PubMed] [Google Scholar]
- 18. Schweitzer L, Geisler C, Pourhassan Met al. . What is the best reference site for a single MRI slice to assess whole-body skeletal muscle and adipose tissue volumes in healthy adults? Am J Clin Nutr 2015;102:58–65. [DOI] [PubMed] [Google Scholar]
- 19. Cespedes Feliciano EM, Popuri K, Cobzas Det al. . Evaluation of automated computed tomography segmentation to assess body composition and mortality associations in cancer patients. J Cachexia Sarcopenia Muscle 2020;11:1258–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Vugt JLA, Levolger S, Gharbharan Aet al. . A comparative study of software programmes for cross-sectional skeletal muscle and adipose tissue measurements on abdominal computed tomography scans of rectal cancer patients: software programmes for body composition measurements on CT. J Cachexia Sarcopenia Muscle 2017;8:285–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weston AD, Korfiatis P, Kline TLet al. . Automated abdominal segmentation of CT scans for body composition analysis using deep learning. Radiology 2019;290:669–79. [DOI] [PubMed] [Google Scholar]
- 22. Park HJ, Shin Y, Park Jet al. . Development and validation of a deep learning system for segmentation of abdominal muscle and fat on computed tomography. Korean J Radiol 2020;21:88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bedrikovetski S, Seow W, Kroon HMet al. . Artificial intelligence for body composition and sarcopenia evaluation on computed tomography: a systematic review and meta-analysis. Eur J Radiol 2022;149:110218. [DOI] [PubMed] [Google Scholar]
- 24. Rizk JG, Streja E, Wenziger Cet al. . Serum creatinine-to-cystatin-C ratio as a potential muscle mass surrogate and racial differences in mortality. J Ren Nutr 2023;33:69–77. [DOI] [PubMed] [Google Scholar]
- 25. Yoshida S, Nakayama Y, Nakayama Jet al. . Assessment of sarcopenia and malnutrition using estimated GFR ratio (eGFRcys/eGFR) in hospitalised adult patients. Clin Nutr ESPEN 2022;48:456–63. [DOI] [PubMed] [Google Scholar]
- 26. Tlemsani C, Durand J-P, Raynard Bet al. . Relationship between the creatinine/cystatin C ratio and muscle mass measured by CT-scan in cancer patients. Clin Nutr ESPEN 2022;51:412–8. [DOI] [PubMed] [Google Scholar]
- 27. Fu EL, Coresh J, Grams MEet al. . Removing race from the CKD-EPI equation and its impact on prognosis in a predominantly White European population. Nephrol Dial Transplant 2023;38:119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Flamant M, Vidal-Petiot E, Metzger Met al. . Performance of GFR estimating equations in African Europeans: basis for a lower race-ethnicity factor than in African Americans. Am J Kidney Dis 2013;62:182–4. [DOI] [PubMed] [Google Scholar]
- 29. Bukabau JB, Yayo E, Gnionsahé Aet al. . Performance of creatinine- or cystatin C-based equations to estimate glomerular filtration rate in sub-Saharan African populations. Kidney Int 2019;95:1181–9. [DOI] [PubMed] [Google Scholar]
- 30. Inker LA, Schmid CH, Tighiouart Het al. . Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med 2012;367:20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pottel H, Delanaye P, Schaeffner Eet al. . Estimating glomerular filtration rate for the full age spectrum from serum creatinine and cystatin C. Nephrol Dial Transplant 2017;32:497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bland JM, Altman DG.. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986;1:307–10. [PubMed] [Google Scholar]
- 33. Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics 1989;45:255–68. [PubMed] [Google Scholar]
- 34. Mosteller RD. Simplified calculation of body-surface area. N Engl J Med 1987;317:1098. [DOI] [PubMed] [Google Scholar]
- 35. Pieters TT, Veldhuis WB, Moeskops Pet al. . Deep learning body-composition analysis of clinically acquired CT-scans estimates creatinine excretion with high accuracy in patients and healthy individuals. Sci Rep 2022;12:9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Macdonald JH, Marcora SM, Jibani Met al. . Bioelectrical impedance can be used to predict muscle mass and hence improve estimation of glomerular filtration rate in non-diabetic patients with chronic kidney disease. Nephrol Dial Transplant 2006;21:3481–7. [DOI] [PubMed] [Google Scholar]
- 37. Cruz-Jentoft AJ, Bahat G, Bauer Jet al. . Sarcopenia: revised European consensus on definition and diagnosis. Age Ageing 2019;48:16–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Albano D, Messina C, Vitale Jet al. . Imaging of sarcopenia: old evidence and new insights. Eur Radiol 2020;30:2199–208. [DOI] [PubMed] [Google Scholar]
- 39. Pickhardt PJ. Value-added opportunistic CT screening: state of the art. Radiology 2022;303:241–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ottosson Frost C, Gille-Johnson P, Blomstrand Eet al. . Cystatin C-based equations for estimating glomerular filtration rate do not require race or sex coefficients. Scand J Clin Lab Invest 2022;82:162–6. [DOI] [PubMed] [Google Scholar]
- 41. Rule AD, Bergstralh EJ, Slezak JMet al. . Glomerular filtration rate estimated by cystatin C among different clinical presentations. Kidney Int 2006;69:399–405. [DOI] [PubMed] [Google Scholar]
- 42. Grubb A, Horio M, Hansson L-Oet al. . Generation of a new cystatin C–based estimating equation for glomerular filtration rate by use of 7 assays standardized to the international calibrator. Clin Chem 2014;60:974–86. [DOI] [PubMed] [Google Scholar]
- 43. Eckfeldt JH, Karger AB, Miller WGet al. . Performance in measurement of serum cystatin C by laboratories participating in the College of American Pathologists 2014 CYS Survey. Arch Pathol Lab Med 2015;139:888–93. [DOI] [PubMed] [Google Scholar]
- 44. Bargnoux A-S, Piéroni L, Cristol J-Pet al. . Multicenter evaluation of cystatin C measurement after assay standardization. Clin Chem 2017;63:833–41. [DOI] [PubMed] [Google Scholar]
- 45. Grubb A, Blirup-Jensen S, Lindström Vet al. . First certified reference material for cystatin C in human serum ERM-DA471/IFCC. Clin Chem Lab Med 2010;48:1619–21. [DOI] [PubMed] [Google Scholar]
- 46. Karger AB, Long T, Inker LAet al. ; College of American Pathologists Accuracy Based Committee and Chemistry Resource Committee . Improved performance in measurement of serum cystatin C by laboratories participating in the College of American Pathologists 2019 CYS Survey. Arch Pathol Lab Med 2022;146:1218–23. [DOI] [PubMed] [Google Scholar]
- 47. Mitch WE, Collier VU, Walser M.. Creatinine metabolism in chronic renal failure. Clin Sci (Lond) 1980;58:327–35. [DOI] [PubMed] [Google Scholar]
- 48. Jones JD, Burnett PC.. Creatinine metabolism in humans with decreased renal function: creatinine deficit. Clin Chem 1974;20:1204–12. [PubMed] [Google Scholar]
- 49. Kurk SA, Stellato RK, Peeters PHMet al. . Trajectory of body mass and skeletal muscle indices and disease progression in metastatic colorectal cancer patients. Am J Clin Nutr 2019;110:1395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.