Fig. 1.
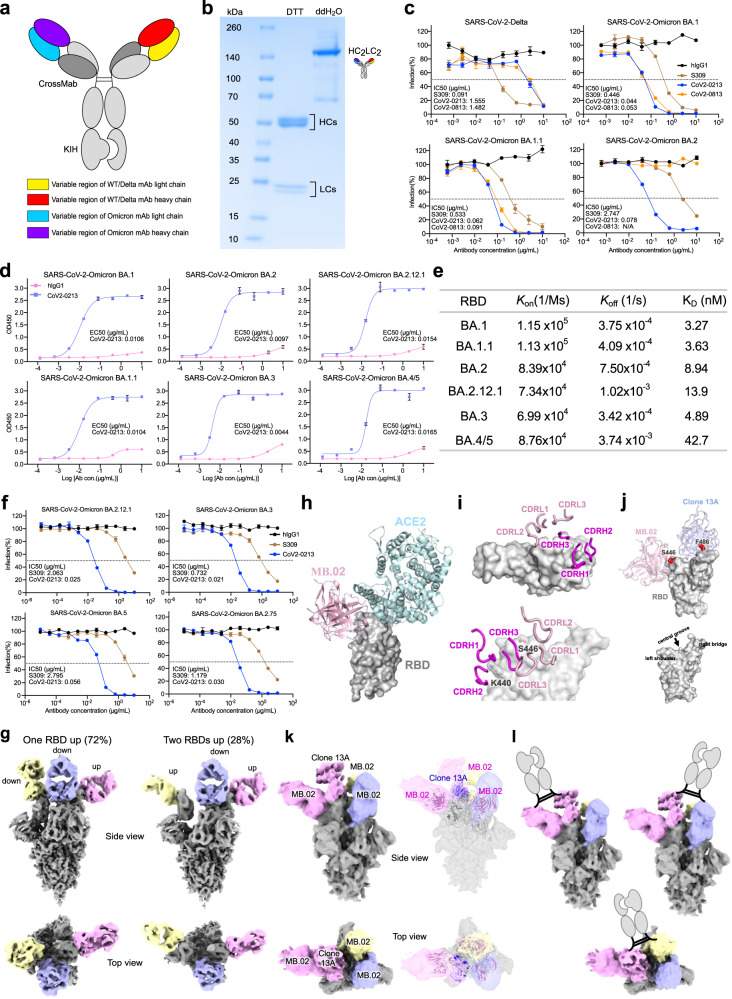
Design, purification, neutralizing activities, and cryo-EM structures of a broadly potent human bispecific antibody CoV2-0213 against circulating Omicron subvariants. a A scheme of bsAb design. Antibody domains are colored according to their architecture. b Coomassie-stained SDS-PAGE analysis of purified bsAb CoV2-0213. The antibody samples are analyzed under reducing conditions (+DTT) and nonreducing conditions (+ddH2O). c Neutralization curves of two lead bsAbs with S309 against the pseudotyped virus of SARS-CoV-2 Delta and Omicron sublineages BA.1, BA.1.1, and BA.2. Serial dilutions of bsAb were added to test its neutralizing activity against indicated pseudotyped virus. The IC50 was determined by log (inhibitor) response of nonlinear regression and is displayed as the mean ± s.e.m. d ELISA binding curves of CoV2-0213 with RBD proteins of SARS-CoV-2 Omicron sublineages BA.1, BA.1.1, BA.2, BA.2.12.1, BA.3, and BA.4/5. The EC50 was determined by log (agonist) response of nonlinear regression and is displayed as the mean ± s.e.m. e The summary statistics of binding affinity of CoV2-0213 to the RBDs of Omicron sublineages as determined by BLI. f Neutralization curves of CoV2-0213 and S309 against pseudotyped virus of Omicron sublineages BA.2.12.1, BA.3, BA.5 and BA.2.75. Serial dilutions of bsAb were added to test its neutralizing activity against indicated pseudovirus. The IC50 was determined by log (inhibitor) response of nonlinear regression and is displayed as the mean ± s.e.m. g Cryo-EM structures of MB.02 Fab fragment in complex with spike trimer in two different conformations, with one RBD up (left) or two RBDs up (right). Fab molecules are shown in different colors, and the spike is shown as dark gray. The corresponding particle distribution of each spike trimer conformation is shown. h Overlay of the structures of human ACE2 (PDB 7wpb) and MB.02 Fab onto the same spike RBD by superimposing the RBD regions. i The binding conformations of the three CDRH and the three CDRL loops of MB.02 Fab on spike RBD. Upper panel, top view; lower panel, side view. The omicron BA.1.1.529 mutation N440K and G446S, which is located at the MB.02 binding interface and inserted between CDRH1 and CDRH2 loops of MB.02 Fab, is shown in stick representation. The nomenclature of the top surface of spike RBD is illustrated in the middle inset. j Overlay of the structures of the humanized clone 2 (Clone 13A) and MB.02 Fab fragments on the same spike RBD. S446 at the MB.02-bound interface of Omicron BA.1/3 spike RBD, and F486 at the Clone 13A-bound interface of WT and Omicron BA.1/2/3 spike RBD, are shown as red spheres. The nomenclature of the top surface of spike RBD is illustrated in the lower panel. k Cryo-EM reconstruction (left panel) and fitted models (right panel) of the CoV2-0213 antibody-bound Omicron BA.5 spike trimer with one RBD up. Fab molecules are shown in different colors, and the spike is shown in dark gray. The up RBD has both MB.02 and Clone 13A Fab fragments bound at distinct interfaces. The other two RBDs in down conformation have the MB.02 Fab bound. The models of the Fab variable domains (MB.02, pink; Clone 13A, blue; RBD, black) are fitted into the 3D reconstruction. l Cartoon illustrations of the possible bivalent binding of one CoV2-0213bsAb onto a single or two adjacent spike RBDs in the same S trimer. The Fc fragment and hinge region of the IgG is shown in the cartoon