Abstract
NUP98 fusions comprise a family of rare recurrent alterations in AML, associated with adverse outcomes. In order to define the underlying biology and clinical implications of this family of fusions, we performed comprehensive transcriptome, epigenome, and immunophenotypic profiling of 2,235 children and young adults with AML and identified 160 NUP98 rearrangements (7.2%), including 108 NUP98-NSD1 (4.8%), 32 NUP98-KDM5A (1.4%) and 20 NUP98-X cases (0.9%) with 13 different fusion partners. Fusion partners defined disease characteristics and biology; patients with NUP98-NSD1 or NUP98-KDM5A had distinct immunophenotypic, transcriptomic, and epigenomic profiles. Unlike the two most prevalent NUP98 fusions, NUP98-X variants are typically not cryptic. Furthermore, NUP98-X cases are associated with WT1 mutations, and have epigenomic profiles that resemble either NUP98-NSD1 or NUP98-KDM5A. Cooperating FLT3-ITD and WT1 mutations define NUP98-NSD1, and chromosome 13 aberrations are highly enriched in NUP98-KDM5A. Importantly, we demonstrate that NUP98 fusions portend dismal overall survival, with the noteworthy exception of patients bearing abnormal chromosome 13 (clinicaltrials gov. Identifiers: NCT00002798, NCT00070174, NCT00372593, NCT01371981).
Introduction
Acute myeloid leukemia (AML) accounts for 15-20% of all pediatric leukemias and is a very heterogeneous disease.1,2 Besides early response to induction treatment assessed by morphology and flow cytometry-based measurable residual disease (MRD), cytogenetic and molecular aberrations are the most important prognostic factors that guide risk group stratification.1,3 Although survival rates of pediatric AML (pAML) patients have improved significantly, over the last decade these have reached a plateau, with long-term survival rates around 70-80%.3,4 A third of all pAML patients relapse, and their outcome is poor.3 In addition, treatment-related toxicity and mortality make intensification of treatment challenging.2,3 Thus, the identification of prognostic subgroups for risk group and treatment stratification is of utmost value to improve treatment and outcomes of specifically high-risk subtypes.2 Due to the very low prevalence of some subgroups, studies to identify these cases can be challenging and therefore require international collaboration.
NUP98 (chromosome 11p15) encodes a nucleoporin protein, which is part of the nuclear pore complex.5 NUP98 was first shown to be fused to HOXA9 in t(7;11) FAB (French-American-British classification) M2 and M4 AML in 1996.6 In the last 20 years, over 30 different partner genes in AML and therapy-related myelodysplastic syndrome have been described.7-10 NUP98 fusion proteins involve the N-terminal portion of NUP98 and the C-terminal portion of the fusion partner.5 These fusion partners consist of homeodomain proteins, which are transcription factors, and non-homeodomain proteins, which are thought to play a role in transcriptional or epigenetic regulation.5 In pAML patients, NUP98 translocations with KDM5A and NSD1 have been most frequently described.11,12 These patients are now notorious for inferior outcome compared to non-NUP98-translocated patients and are treated as high-risk patients in most current treatment protocols.9,13 However, NUP98 translocations with other partners, here called NUP98-X, are rare, and their prognostic relevance is unknown; consequently, there is a necessity to define the optimal risk stratification and treatment strategy for these patients. Here, we present the molecular and clinical characteristics of NUP98-translocated pAML patients within four consecutive Children’s Oncology Group (COG) trials and an International Berlin-Frankfurt-Münster AML study group (I-BFM AML SG) collaboration. We aim to define the clinical relevance for all NUP98 translocations with cooperating mutations and copy number variants.
Methods
Patient samples
Patients enrolled in the COG trials CCG-2961, AAML03P1, AAML0531 and AAML1031 were eligible for this study. Details of these studies have been previously described.14-17 In total, 3,493 patients were included in these studies, of which 2,235 were eligible for inclusion due to availability of comprehensive NUP98 fusion, molecular, and clinical data (Online Supplementary Figure 1; Online Supplementary Tables S1 and S2). For the remaining patients, these data were unavailable. In addition, we sent out an I-BFM AML SG proposal to include pediatric AML patients with a NUP98-X translocation from other study groups. Consent, in accordance with the Declaration of Helsinki, was obtained from all study participants. The Fred Hutchinson Cancer Research Center Institutional Review Board and the COG Myeloid Biology Committee approved and oversaw the conduct of this study. Adult AML patients from the Beat AML study, The Cancer Genome Atlas AML (TCGA LAML), and Southwestern Oncology Group (SWOG) AML studies were included as comparators for NUP98 fusion analysis and details were reported accordingly in references.18-23
Screening of NUP98 fusions
The NUP98 fusions were detected by either karyotype or combined fusion detection algorithms STAR-fusion v1.8.1, TransAbyss v1.4.10, and CICERO v0.1.824-2 6 completed on RNA sequencing (RNA-seq). For differences in detection methods per COG trial, see the Online Supplementary Appendix. The majority (94%) of NUP98-translocated patients had RNA-seq evidence of their fusion. STAR-fusion was run using default parameters with the premade GRCh37 resource library with Gencode v19 annotations (h ttps://data.broadinstitute.org/Trinity/CTAT_RESOURCE_LIB/ ). The TransAbyss software was executed with the GRCh37-lite reference genome with the following parameters included: fusion breakpoint reads ≥1, flanking pairs and spanning reads ≥2 counts. CICERO fusion detection was performed with default parameters with GRCh37-lite. Fusions detected computationally were verified using Fusion Inspector v.1.8.1 (Broad Institute, Cambridge, MA) and visualized on IGV27-30 and BAMBINO.31 Beat AML (n=440) and SWOG AML (n=206) transcriptome sequence reads were analyzed using STAR-fusion v1.8.1 with the same reference resource library and parameters as above.24 TCGA LAML (n=179) RNA-seq fusion data were downloaded from supplementary materials.19
Statistical methods
Data were current as of March 31, 2019. The Kaplan-Meier method was used to estimate overall survival (OS, defined as time from study entry to death) and event-free survival (EFS, time from study entry until failure to achieve complete remission [CR] during induction, relapse, or death). Relapse risk (RR) was calculated by cumulative incidence methods defined as time from the end of induction I for patients in CR to relapse or death, where deaths without a relapse were considered competing events. Patients who withdrew from therapy due to relapse, persistent central nervous system (CNS) disease, or refractory disease with >20% bone marrow blasts by the end of induction I were defined as induction I failures. MRD was defined at the end of course one using flow cytometry with a cut-off of 0.1% detection of disease. The I-BFM patients were excluded from survival analyses due to variation in study groups and treatment protocols.
Results
Clinical characteristics
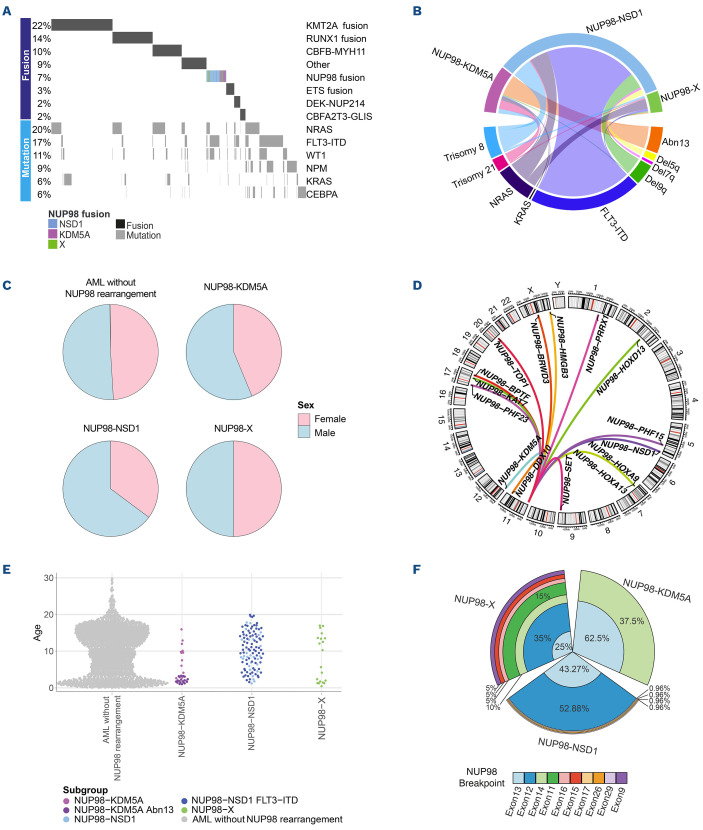
Between 1995 and 2017, 3,493 AML patients were treated on consecutive COG trials CCG-2961, AAML03P1, AAML0531, and AAML1031, of which 2,235 were eligible for comprehensive outcome (see Methods) and cytomolecular association analyses. Within this cohort, 160 patients (7.2%) with a NUP98 translocation were identified (Figure 1A); the remaining 2,075 patients were included as a reference cohort. In addition, six NUP98-X cases were included via the I-BFM AML SG, demonstrating that while rare, NUP98-X cases are present in multiple cohorts of patients. However, to prevent bias due to confounding variables, such as differences in study groups, fusion identification methods and treatment protocols, these patients were excluded from further analyses. Characteristics of all NUP98-X patients are depicted in the Online Supplementary Table S3.
The most common NUP98 translocations were NUP98-NSD1 (n=108) and NUP98-KDM5A (n=32; Online Supplementary Figure S2A). Furthermore, we identified 20 patients with 13 different NUP98 translocation partners, including HOXA9 (n=4), HOXD13 (n=3), PHF15 (n=2), PHF23 (n=2) and single cases of BPTF, BRWD3, DDX10, HMGB3, HOXA13, KAT7, PRRX1, SET, and TOP1 (Figure 1B). Interestingly, contrasting the cryptic NUP98-NSD1 and NUP98-KDM5A fusions, an overwhelming majority of NUP98-X fusions were detectable as karyotypic variants with 17 of 20 (85%) having gross alterations by g-banding cytogenetics involving chr11p15.
Initial comparison of the NUP98 fusion cohort to the reference patients demonstrated a significant sex bias in NUP98 cases, with 61.3% being male versus 38.8% female (P=0.012) (Figure 1C). In particular, the NUP98-NSD1 cohort contained 64.8% male versus 35.2% female patients. Additionally, NUP98 fusions were enriched in children aged 3-10 years old (35.6%; P=0.005). Clinical characteristics for NUP98-trans-located subgroups are summarized in Figure 1C-E and the Online Supplementary Table S1. In NUP98-NSD1-translocated patients, white blood cells and blast cell counts were both significantly higher, while in NUP98-KDM5A patients a reverse trend was seen. Classification by conventional cytomolecular stratification schemas, as previously described,17 revealed that 39% of NUP98-NSD1 patients had been classified as standard-risk (SR) and 61% as high-risk (HR). In contrast, most NUP98-KDM5A patients (97%) and NUP98-X patients (95%) were classified as SR.
Comparison of NUP98 translocations with age at diagnosis based on fusion partners (Figure 1C) showed that NUP98-NSD1 cases had a median age of 10.2 years (reference cohort 10.0; P=0.228), whereas NUP98-KDM5A cases had a median age of 2.7 years (P<0.001). NUP98-X patients showed a median age of 7.9 years (P=0.30) with a bi-modal distribution; 40% of the patients were under 3 years and 50% over 5 years with no patients over 18 years old (Online Supplementary Figure S2B, C). Almost all NUP98-X patients with homeobox fusion partners (n=9) were over 3 years old (8/9), with one exception (NUP98-HOXD13; P=0.025).
From Beat AML, TCGA LAML and SWOG, 825 adult AML cases were screened for NUP98 fusions by RNA-seq fusion detection algorithms. Zero NUP98-KDM5A, 11 (1.3%) NUP98-NSD1, and two (0.2%) NUP98-X fusions were identified (NUP98-TOP1 and NUP98-RAP1GDS1). These results demonstrate that NUP98 rearrangements are less common, but still present, in older adult AML patients (13/825, 1.6%) compared to pediatric and young adult AML patients (160/2,235, 7.2%; P<0.001; Online Supplementary Figure S2D)9.
Implications of variation of fusion junction
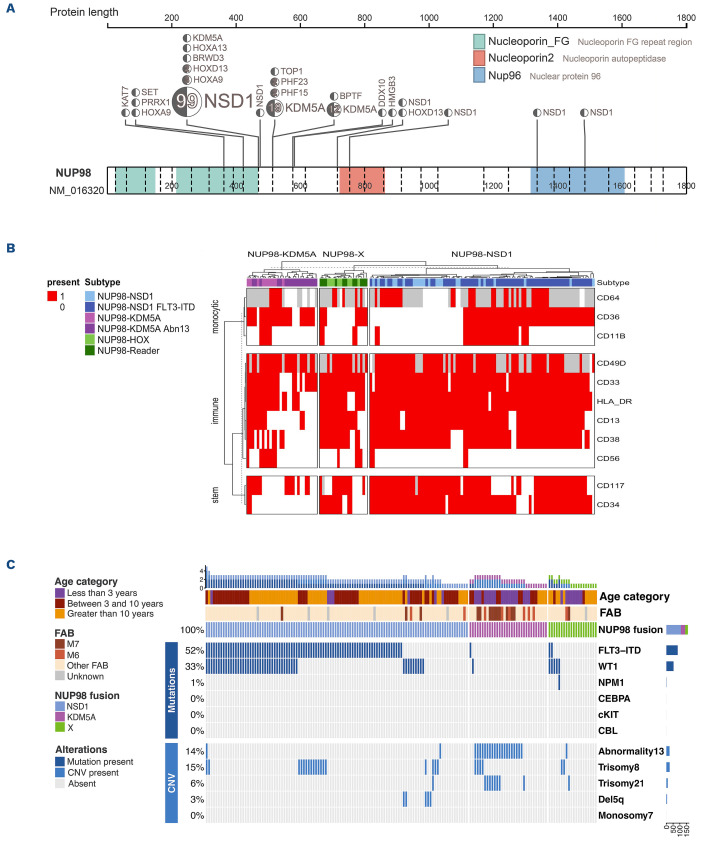
Analysis of NUP98 fusion breakpoints by RNA-seq revealed a high diversity of NUP98 exon junctions. Nearly 85% of NUP98 fusions had a breakpoint junction in exon 12 (39.7%) or 13 (44.9%), while the remaining breakpoints occurred in various positions from exon 11 to exon 29 (Online Supplementary Figure S2E). Exon junctions correlated with the fusion partner, and NUP98-NSD1 fusions primarily had exon 12 (52.9%) and 13 (43.23%) junctions (Figure 1F). However, exon 14 breakpoints were almost uniformly restricted to NUP98-KDM5A compared to other NUP98 fusions (P<0.001). NUP98-X cases showed a larger variability in NUP98 exon breakpoints (Online Supplementary Table S4; Online Supplementary Figure S3). NUP98 homeobox gene fusions were enriched in exon 12 breakpoints (6/9, 66%), while PHF15 (n=2), PHF23 (n=2), and TOP1 partners had exon 13 breakpoints. Besides the commonly included nucleoporin FG repeat domains of the NUP98 protein, a minority of cases (n=3) included a larger portion of the protein with nucleoporin autopeptidase or, additionally, the Nup96 domains (Figure 2A).
Immunophenotypes
NUP98 fusions were previously reported to be associated with erythroid and megakaryocytic phenotypes.10,32 Upon morphology, we identified that only NUP98-KDM5A fusions were more often associated with the FAB M6/M7 category compared to the reference cohort (46.9% vs. 5.5%; P<0.001). Additionally, the immunophenotype of NUP98 fusions was examined using multidimensional flow cytometry.33 NUP98-NSD1 patients expressed early progenitor markers such as CD34 and CD117 (Figure 2B; Online Supplementary Figure S4). Patients harboring NUP98-NSD1 and FLT3 internal tandem duplication (-ITD) retained the immature markers but also showed evidence of monocytic maturation compared to NUP98-NSD1 without FLT3-ITD, as demonstrated by expression of CD11b (84%), CD36 (55%) and CD64 (71%) (Online Supplementary Table S6). Nevertheless, NUP98-NSD1 associated phenotypes were not as specific, or consistent, as seen in NUP98-KDM5A. The NUP98-KDM5A immunophenotype corresponded to mega-karyocytic maturation, with at least partial expression of CD36, and absence of pluripotent markers CD34 and CD123. Notably, NUP98-KDM5A patients showed clusters that associated with co-occurrence of abnormal chromosome 13q (Figure 2C), demonstrating that these subsets of patients display a unique immunophenotype. Finally, NUP98-X fusions lacked consistent immunophenotype, aside from the majority expressing markers of early progenitors.
Figure 1.
Clinical characteristics of patients with and without NUP98 translocations. (A) Oncoprint depicting the major drivers of pediatric acute myeloid leukemia (AML) patients. (B) Circos plot depicting commonly co-occurring mutations and cytogenetic abnormalities in NUP98-translocated pediatric AML patients. (C) Pie charts depicting the sex divisions of patients in the NUP98-translocated AML subgroups. (D) Circos plot representing different fusion partner genes of NUP98-X translocations in pediatric AML patients. (E) Age distribution of AML patients. (F) Barchart (polar axis) illustrating the prevalence of NUP98 exon junctions in NUP98-translocated AML. The Figure legend is ordered by decreasing NUP98 exon prevalence in the NUP98 fusion-positive cohort.
Cooperating karyotypic and molecular variants
Diagnostic specimens were evaluated for common translocations, chromosomal aberrations and common mutations, namely FLT3-ITD, WT1, NPM1, CEBPA, KIT and CBL mutations (Figures 1A and 2C). We confirmed the well-known enrichment of FLT3-ITD (74%) and WT1 mutations (42%) in the NUP98-NSD1 cohort.5 Almost half of NUP98-NSD1 patients with FLT3-ITD also harbored a WT1 mutation, indicating triple positivity for adverse outcome variants in AML.34 NUP98-NSD1 patients also had a significant association with trisomy 8 (18.8%) compared to the reference cohort (5.3%; P<0.001). NUP98-KDM5A displayed a paucity of cooperating mutations. NUP98-X patients showed a higher prevalence of WT1 mutations compared to the reference cohort (25% vs. 9.6%; P=0.039), associated with a higher age at diagnosis (median age 16.3 vs. 2.3 years; P=0.032). We identified a notably high correlation of NUP98-KDM5A with chromosome 13 (chr13) structural variants, including del(13q), monosomy 13 and chr13 translocations. Abnormal chr13 (NUP98-KDM5A/13abn) was identified in 19 NUP98-KDM5A patients (63.3% vs. 2.3% in the reference cohort; P<0.001). NUP98-KDM5A/13abn were significantly younger than NUP98-KDM5A/13normal patients (median age 1.8 vs. 9.6 years; P<0.001). Thirteen NUP98-KDM5A patients (43.3%) patients harbored del(13q) versus one NUP98-X patient (NUP98-SET). Monosomy 13 (2/32) and translocation 13 (4/32) occurred less frequently in NUP98-KDM5A and were not found in NUP98-NSD1 or NUP98-X (Online Supplementary Table S5).
The majority of del(13q) in NUP98-translocated cases (92%) began at band 13q12; the deletions ranged from 8 Mb to 59.5 Mb to the entire chromosome. The minimal commonly deleted segment was del(13)(q14.2q14.3), containing the RB1 tumor suppressor gene (Online Supplementary Figure S5).
RB1 loss has been previously reported in patients with NUP98-KDM5A;10 however, we demonstrated a much larger region of copy number alterations, including numerous additional genes. Of NUP98-KDM5A/13abn patients, 84% (16/19) had NUP98 exon 13 breakpoints, suggesting that specific NUP98 exon breakpoints in the fusion transcript may be linked to the presence of additional cytogenetic abnormalities. Finally, all ten acute megakaryoblastic leukemia (AMKL; FAB M7) NUP98-KDM5A cases with karyotype data available had chr13 alterations compared to three AMKL cases without NUP98 fusions (P<0.001).
Gene expression profiling
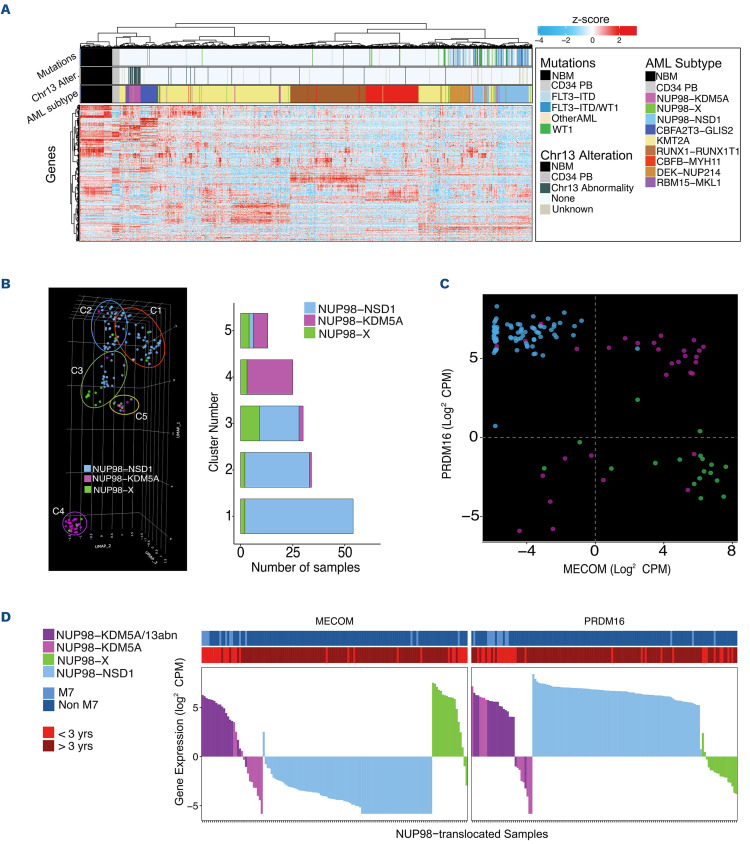
Unsupervised hierarchal clustering of gene expression in NUP98-translocated patients and a reference cohort of known fusions including KMT2A, CBFB-MYH11, RUNX1-RUNX1T1 and DEK-NUP214, as well as 84 healthy controls (n=988), revealed that the majority of NUP98-NSD1 (n=104), NUP98-KDM5A (n=32), and the reference cohort cluster by fusion identity, while no uniform clustering of NUP98-X was observed (n=20) (Figure 3A). In order to further understand transcriptional similarities and differences between the diverse NUP98 fusions, uniform manifold approximation and projection (UMAP) was completed on the NUP98-translocated patients’ gene expression data (n=156). The Leiden algorithm35 identified five transcriptional clusters. NUP98-NSD1 patients clustered together, clearly separated from the majority of NUP98-KDM5A patients (Figure 3B). NUP98-X patients were dispersed, clustering more closely with NUP98-NSD1 cases. The largest proportion of NUP98-X clustering together included seven homeobox and both PHF23 partners, suggesting transcriptional similarities between NUP98-HOX fusions (cluster C3). The next cluster most associated with NUP98-X (cluster C5) included the majority of non-AMKL NUP98-KDM5A cases.
UMAP revealed segregation based on an AMKL and age-based signature (cluster C4), which embodied 78.6% of AMKL NUP98-translocated patients, all 3 years old or younger. The cluster primarily contained NUP98-KDM5A (22/32) cases and was enriched in NUP98 exon 13 breakpoints. NUP98-X patients in C4 included single cases of NUP98-SET with del(13q), NUP98-BPTF with AMKL morphology, and NUP98-DDX10. Additionally, C4 included all NUP98-KDM5A/13abn cases (Online Supplementary Figure S6A), separating NUP98-KDM5A with and without chr13 abnormalities. Conversely, in a separate UMAP including heterogenous pAML fusions (N=1,482), this abn13-based clustering was not observed for non-NUP98-translocated subtypes (Online Supplementary Figure S6B).
Differential expression analysis compared NUP98-X directly to NUP98-NSD1 and NUP98-KDM5A individually. Expression of MECOM and PRDM16, known prognostic markers in adult and pediatric AML,36 effectively separated the NUP98 subgroups (Figure 3C). Interestingly, about two-thirds of NUP98-KDM5A highly expressed both genes, while NUP98-X and NUP98-NSD1 almost exclusively over-expressed one or the other (Figure 3D). NUP98-KDM5A patients with low MECOM and low PRDM16 expression almost uniformly lacked chr13 alterations. Additionally, NUP98-KDM5A/13abn patients had reduced expression of genes in the involved area, including RB1 (P<0.001), DLEU7 and SPRYD7 (Online Supplementary Figure S6C).
Figure 2.
Cytogenetics of A/l/P9S-translocated pediatric acute myeloid leukemia. (A) Locations of breakpoints across the NUP98 gene for all NUP98-translocated acute myeloid leukemia (AML). (B) Oncoprint depicting additional copy number variations (CNV) and mutations in NUP98-translocated patients. (C) Heatmap depicting the presence and absence of flow-cytometry immunophenotype markers in NUP98-translocated AML groups. NUP98-HOX-like fusions include fusion partners HOXA9, HOXA13, HOXD13, and PRRX1. NUP98-Reader-like fusions include fusion partners BPTF, BRWD3, DDX10, HMGB3, KAT7, PHF15, SET, and TOP1.
Figure 3.
Expression patern of pediatric acute myeloid leukemia with various NUP98 translocations. (A) Unsupervised hierarchical clustering by gene expression including heterogenous pediatric acute myeloid leukemia (AML) subtypes, NUP98-translocated subgroups, normal healthy bone marrows (NBM) and CD34+ peripheral blood cells (CD34 PB). Annotation bars show AML subtype and co-occurring mutations. (B) Uniform manifold approximation and projection (UMAP) of gene expression, followed by Leiden clustering, for NUP98-translocated pediatric AML samples identifies five different transcriptional clusters. NUP98 fusions are indicated in different colors: NUP98-KDM5A in purple, NUP98-NSD1 in blue, and NUP98-X in green. (C) Expression of MECOM and PRDM16 genes in different subgroups of NUP98-translocated pediatric leukemia. Same identification colors for NUP98 fusions as in (B) are used. (D) Expression of stemness marker genes in all NUP98-translocated samples. Top bars represent French-American-British (FAB) classification and age category.
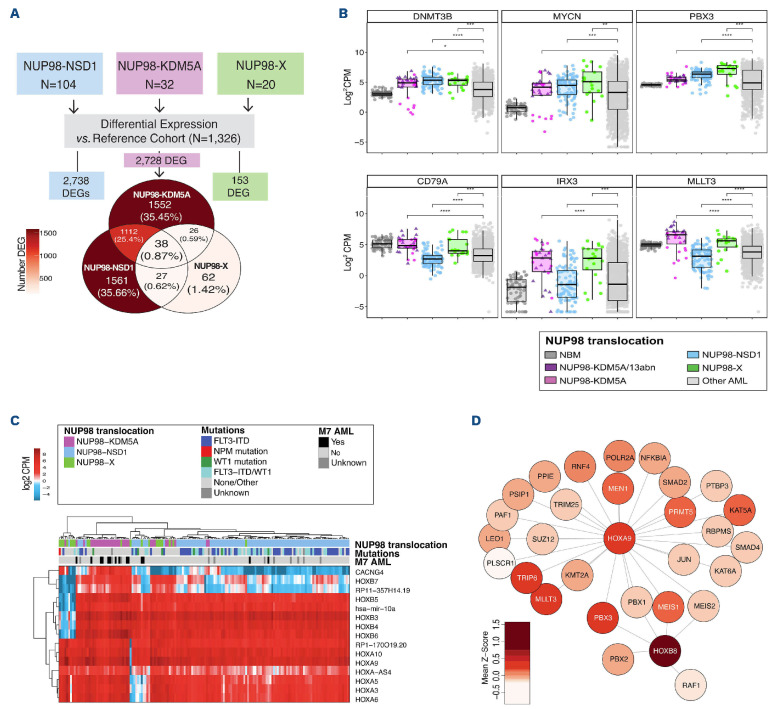
We attempted to identify transcriptional signatures that might be shared between all NUP98-translocated cases and performed differential expression analysis comparing each group (NUP98-X, NUP98-NSD1, and NUP98-KDM5A) independently to the reference cohort (n=1,326) (Figure 4A). This analysis confirmed high inter-patient variability of NUP98-X fusions (Online Supplementary Figure S7). Gene expression profiling revealed 27 differentially expressed genes (DEG) exclusively shared between NUP98-X and NUP98-NSD1, including upregulation of DNMT3B, MYCN, and PBX3 (Figure 4B). Within NUP98-KDM5A, a bimodal expression pattern of DNMT3B and MYCN was related to chr13 alterations, where cases lacking chr13 aberrations had decreased expression. NUP98-X and NUP98-KDM5A exclusively shared 26 dysregulated genes, including overexpression of MLLT3, IRX3, and CD79a.
The NUP98-translocated cohort had 38 DEG in common, including upregulation of numerous HOX genes. Among these 38 genes, 15 were also dysregulated in NUP98-translocated cohorts compared to healthy bone marrow samples. This minimal set of 15 genes strongly implicated dysregulation at the HOX loci; these targets include HOXA (chr7p15), HOXB (chr17q21), hsa-mir-10a (chr17q21), and CACNG4 (chr17q24) transcripts (Figure 4C). NUP98-X cases expressed HOXA/B genes regardless of their fusion partner, and 60% (12/20) expressed both HOXA/B while the remaining third primarily overexpressed the HOXA cluster (Online Supplementary Figure S8). Overexpression of HOX genes and hsa-mir-10a was previously reported in NUP98-KDM5A and NUP98-NSD1 and is now shown to be a common feature of NUP98 translocations.9,13
Single-sample gene-set enrichment analysis (ssGSEA) addresses the inherent variability within diverse NUP98 fusions and was performed to investigate alterations in the expression of down-stream targets of hsa-mir-10a and HOX transcription factors.37 NUP98-translocated subgroups had significantly lower enrichment scores of miR-10a-3p and miR-10a-5p/miR-10b-5p target genes, an indication of negative regulation, compared to normal bone marrow samples (P<0.001). Investigation of HOX transcription factor (TF) pathways by ssGSEA, revealed enrichment in HOXB8 molecular interactions (adj.P<0.008). The HOXB8 pathway included well known HOX transcriptional co-factors MEIS1, MEIS2, PBX1, PBX3, PBX3,38 and the proto-oncogene RAF1. NUP98-X and NUP98-KDM5A exhibited a positive enrichment of HOXA9 interacting partners (P<0.001; Figure 4D). Additionally, we employed RCIS-Target to identify TF motifs enriched in the overexpressed genes (fold-change >2.0) for each NUP98-translocated cohort (Online Supplementary Table S7). This revealed a transcriptional network in NUP98-KDM5A with GATA1 and GATA2 both highly upregulated compared to the reference cohort, and their downstream target genes concomitantly overexpressed, with concurrent down-regulation of ERG, which is known to have an inverse relationship with GATA expression.39
DNA methylation profiling
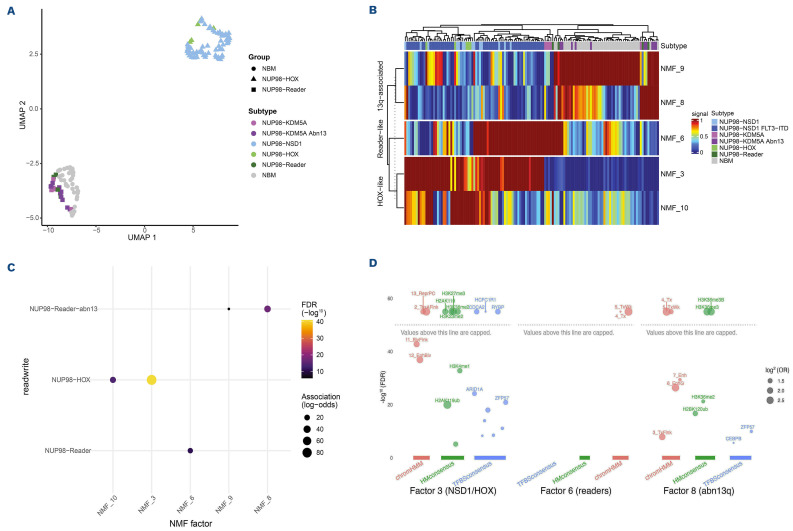
We analyzed DNA methylation data from 334,934 CpG probes. We then performed dimensionality reduction using non-negative matrix factorization (NMF) and used UMAP to determine how the variation in DNA methylation associates with NUP98 fusion groups and normal bone marrow (NBM). We found that NUP98 fusion groups cluster together (Figure 5A). Specifically, the HOX-activating fusions (NSD1, HOX, and PRRX1) form a unique cluster, and also the fusion partners with reader-like functions (BPTF, BRWD3, DDX10, HMGB3, KAT7, PHF15, PHF23, SET, and TOP1) cluster together. The reader-like fusions also cluster more closely to NBM. By performing unsupervised clustering of the NMF factors that associate with each group, we found that the NUP98-HOX-like group clusters distinctly from the NUP98-readers and NBM, further illustrating that NUP98 fusions differ in methylation profiles (Figure 5B).
We further analyzed the NMF factors that associate significantly with the NUP98-HOX, NUP98-Reader, and NUP98-Reader plus abn13 groups (Figure 5C). In order to identify the defining characteristics within each of these factors, we performed enrichment analyses against chromatin states, histone marks, and transcription factor binding sites (Figure 5D). The NUP98-HOX group enrichments in NMF 3 indicate that these fusions lead to Polycomb-mediated hypermethylation at actively transcribed genes, evidenced by H2AK119ub, H3K23me2, H3K36me2/3, and H3K27me3 enrichment at binding motifs for RYBP (a subunit of Polycomb repressor complex 1). This likely occurs because H3K36me2/3 increases throughout the HOXA/B clusters and at HOX targets, while H3K27me3 peaks disappear as hyperactive NSD1 displaces PRC1/2 from the HOX clusters. This may result in reducing expressing potential and arresting cellular differentiation, which often coincides with loss of imprinting, as evidenced by dual enrichment for H3K27me3 and H3K36me3, as well as transcription factor binding site enrichment for ZFP57, the master regulator of genomic imprinting control regions. The enrichment of NMF 6 suggests that NUP98-Reader fusions likely lead to localization of transcriptional condensates at already highly expressed developmental genes, leading to an enrichment of DNA hypermethylation in transcribed exons.
The enrichment of NMF 8 suggests that abnormal chr13 cases, all of which are NUP98-Reader fusions, show additional hypermethylation of actively expressed gene bodies, evidenced by enrichment for H3K36me3 and H2BK120ub, along with loss of imprinting (though far less pronounced than in NUP98-HOX fusions), which is evidenced by an enrichment for ZFP57 binding sites.
Clinical outcome and prognostic relevance
We evaluated the impact of NUP98 translocations on response to initial induction therapy. Overall, the morphologic CR rate after course one for the NUP98 fusion cohort was 50% versus 78% for the reference cohort (P<0.001). NUP98-NSD1 patients had a significantly lower CR rate of 38% (P<0.001) compared the reference cohort, while NUP98-
Figure 4.
Differential expression of all NUP98-translocated pediatric acute myeloid leukemia patients. (A) Schematic of differential expression analyses completed for NUP98-translocated samples. The overlap of differentially expressed genes (DEG) identified in each NUP98 cohort is represented in the Venn diagram. (B) DEG between NUP98-translocated AML groups compared to the reference cohort and normal bone marrow (NBM) were identified. Subsets of dysregulated genes were commonly identified in both NUP98-X and NUP98-NSD1 (upper panel) or were identified as shared between NUP98-X and NUP98-KDM5A (lower panel). (C) Commonly DEG found in all three NUP98-translocated pediatric acute myeloid leukemia (AML) subgroups. (D) Mean expression (Z-score transformed) of HOXA9 and HOXB8 interacting partners in NUP98-X translocated AML. The darker shades of red indicate higher expression in the NUP98-X cohort. CPM: counts per million.
Figure 5.
DNA methylation of pediatric acute myeloid leukemia patients with NUP98 translocations. (A) Uniform manifold approximation and projection (UMAP) of DNA methylation data in A/L^PSS-transLocated acute myeloid leukemia (AML) subgroups compared to the reference cohort and normal bone marrow (NBM). (B) Heatmap of non-negative matrix factorizations (NMF) of DNA methylation data. The NMF factors are those that were significantly associated with NUP98 translocation AML subgroups. (C) NMF factor associations of DNA methylation with NUP98-HOX-[\ke fusions (NUP98-NSD1, NUP98-HOX, and NUP98-PRRX1) and NUP98-Reader-like fusions (NUP98-KDM5A, NUP98- BPTF, NUP98-BRWD3, NUP98-DDX10, NUP98-KAT7, NUP98- PHF15, NUP98-SET, and NUP98-TOP1) with or without a co-occuring abnormal chromosome 3 (chr3). (D) NMF factor enrichments of DNA methylation for chromatin states, chromatin marks, and transcription factor binding sites. Factor 3 is enriched in NUP98-HOX-Like fusions, factor 6 is enriched in NUP98-Reader-like fusions, and factor 8 is enriched in NUP98-Reader-like fusions with an abnormal chr13. FDR: false discovery rate; OR: odds ratio; ReprPC: repressed PolyComb; TssAFlank: flanking active TSS, BivFlank: flanking bivalent; TSS/Enh, EnhBiv: bivalent enhancer; TxWk: weak transcription; Tx: Strong transcription; Enh: enhancers; EnhG: genie enhancers.
Figure 6.
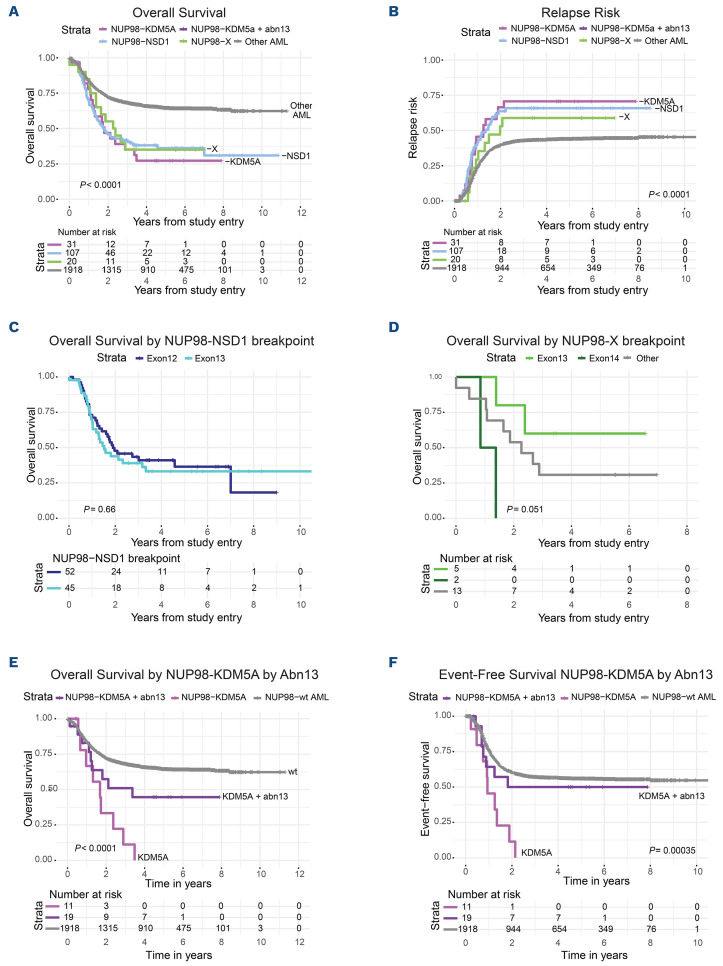
Survival of pediatric acute myeloid leukemia patients with NUP98 translocations. Kaplan Meier estimates of (A) overall survival (OS) and (B) relapse risk (RR) of pediatric NUP98-translocated acute myeloid leukemia (AML) patients with different translocation partners compared to a reference cohort without NUP98 fusions. OS of (C) NUP98-NSD1 and (D) NUP98-X, when divided by NUP98 fusion exon breakpoint. Outcome was also examined for (E) OS and (F) event-free survival (EFS) of NUP98-KDM5A subgroups by chromosome 13 (chr13) status (monosomy 13, del(13q), translocation 13). Abn3: abnormal chr 3.
KDM5A and NUP98-X had CR rates of 81% (P=0.729) and 65% (P=0.176), respectively. NUP98-NSD1 and NUP98-KDM5A patients had significantly higher evidence of MRD (73%; P<0.001, and 52%; P=0.005, respectively), while this was similar to the reference cohort in NUP98-X (22% vs. 27%; P=0.793).
The 5-year OS for the NUP98 fusion cohort was 35% versus 64% for the reference group (P<0.001). NUP98-NSD1 patients had inferior OS (36% vs. 64%, P<0.001) and event-free survival (EFS) (17% vs. 47%; P<0.001) compared to the reference (Online Supplementary Table S1; Figure 6A, B). Similarly, adverse outcomes for NUP98-KDM5A were observed for OS (30%; P<0.001) and EFS (25%; P=0.01). NUP98-NSD1 and NUP98-KDM5A cases showed a significantly higher 5-year relapse risk (RR) of 64% (P=0.001) and 68% (P=0.010) respectively, compared to the reference cohort (42%) (Figure 6B). NUP98-X displayed a similar inferior OS (35%; P=0.009); however, EFS (35%; P=0.333) and RR (69%; P=0.071) differences did not reach significant difference. Response to treatment in NUP98-translocated subgroups, examined by disease-free survival (DFS) estimates 5 years after induction one, was lower compared to the reference cohort (27% vs. 52%; P<0.001). This held true for all subsets; NUP98-NSD1 (28%; P<0.001), NUP98-KDM5A (28%; P=0.012) and NUP98-X (23%; P=0.044) (Online Supplementary Figure S9A).
Multivariable cox regression analyses were performed to adjust for cytomolecular risk groups, white blood cells, and different NUP98-translocated subgroups (Online Supplementary Table S8). After correction, significantly inferior OS (hazard ratio [HR]=1.463; 95% confidence interval [CI]: 1.1-1.94; P=0.009), EFS (HR=2.032; 95% CI: 1.59-2.59; P<0.001) and RR (HR=1.743; 95% CI: 1.1-2.76; P=0.018) were observed in NUP98-NSD1 patients compared to the reference group. Also, NUP98-KDM5A (HR=1.825; 95% CI: 1.13-2.96; P=0.015) and NUP98-X patients (HR=1.75; 95% CI: 1.01-3.04; P=0.046) showed poor OS, without significant differences in EFS and RR.
We examined outcomes corresponding to fusion exon junctions (Figure 6C, D; Online Supplementary Figure 9A, B). There were no significant differences in outcome for NUP98-NSD1 or NUP98-X by exon junction, though a trend toward improved outcomes was observed for NUP98-X exon 13 breakpoints. NUP98-KDM5A patients with exon 13 junctions (n=19) had an OS of 51% compared to 0% for exon 14 breakpoints (n=12; P=0.011) with corresponding EFS (40% vs. 0%, respectively; P=0.174). Due to high concurrence of chr13 alterations with exon 13 junctions, a similar trend was observed in NUP98-KDM5A/13abn compared to NUP98-KDM5A/13normal patients (EFS 45% vs. 0%; P=0.052). NUP98-KDM5A patients had a worse prognosis compared to the reference cohort without NUP98 fusions regardless of chr13 alterations; however, the presence of chr13 alterations within the NUP98-KDM5A group was associated with increased OS and EFS (Figure 6E, F).
Discussion
NUP98-translocated pAML has emerged as a distinct but heterogeneous group, and a comprehensive study defining varied fusion partners, phenotypes, transcript subclasses and outcomes was still lacking. Incorporation of genome, tran-scriptome, methylation, and clinical data from several large pediatric and adult AML studies provided deep insight into this family of fusions. Our study demonstrates that the underlying biology of NUP98-translocated AML is defined by the fusion partner. Furthermore, although fusions involving NSD1 and KDM5A are cryptic, an overwhelming majority of NUP98-X fusions can be identified by conventional karyotype, facilitating identification at diagnosis.
Importantly, we identified a significant overlap of cooperating lesions including mutations (FLT3, WT1) and karyotypic alterations (trisomy 8, del13q). We confirmed prior observation of substantial enrichment of FLT3-ITD in NUP98-NSD1 patients (80%). This extreme prevalence, and NUP98-NSD1 preceding FLT3-ITD, suggest a causal relationship; this intriguing hypothesis is being studied in our laboratory.
Recently, exon usage and fusion junctions were shown to have clinical and biological implications; patients with a CBFB-MYH11 fusion with the common exon 5/33 breakpoint had significantly inferior EFS than those with less common fusion junctions.40 We here demonstrated that patients with NUP98-KDM5A with exon 13 involvement had a more favorable prognosis. However, the strong association of exon 13 usage with chr13 alterations in NUP98-KDM5A patients makes it difficult to discern which of these factors is underlying this outcome difference (Figure 6; Online Supplementary Figure S9). The difference that we discovered in prognosis may suggest that NUP98-KDM5A cases with or without exon 13 breakpoints and chr13 abnormalities could be divided into different subgroups. The observation that NUP98-KDM5A/abn13 patients have a more favorable prognosis potentially affects treatment stratification of these patients in future. Furthermore, these findings provide a rationale that future studies must go beyond simple defining the presence or absence of a fusion and investigate specific exon usage, inclusion/exclusion of critical functional domains, and functionality of the oncoprotein.
Transcriptome profiling further defined functional classifications of NUP98 fusions. Expression of PRDM16 and MECOM could clearly segregate NUP98-translocated subsets. PRDM16 and MECOM encode H3K9-monoDmethyltransferases that are important in the maintenance of heterochromatin integrity and are selectively expressed in hematopoietic stem cells (HSC)36 and linked to oncogenic transformation;41 their deregulation could play a role in leukemogenesis of NUP98 translocations. Gene expression profiling also revealed distinct expression networks defined by translocation partner and cooperating mutations/alterations. Not only did NUP98-KDM5A patients cluster based on abn13, bimodal expression of DNMT3B, MYCN, MECOM, and PRDM16 was associated with abn13. Interestingly, where PRDM16 is a poor prognostic factor in AML,36 high expression was associated with NUP98-KDM5A/abn13, which had a better prognosis in our cohort. This association may indicate different molecular pathways underlying leukemogenesis within NUP98-KDM5A, where NUP98-KDM5A/13abn may have more immature HSC-like features.
Regardless of fusion partner, NUP98 translocations shared overexpression of HOXA/B genes. The translocation partners KDM5A and PHF23 contain PHD protein domains, which function in histone methylation and nucleosome remodelling.25 The HOX cluster was shown to be in a locked, transcriptionally active position due to the H3K4me3-binding PHD-domain when fused to NUP9825 and this may be extendable to the leukemogenic ability of PHF15 and BPTF, which retain their PHD finger. Translationally, upregulation of the HOXA cluster indicates a potential therapeutic role of menin-inhibitors in NUP98-translocated AML, as has been recently shown in mice42 and in vitro studies of primary pAML samples.43
Chromatin modifiers, such as NSD1 and KDM5A, are frequently the targets of oncogenic fusions in pediatric disease.44 DNA methylation profiling suggests that rare and diverse NUP98-X fusions share one of the two key mechanisms to promote leukemogenesis: either by activating the HOX genes and their targets and promoting loss of genomic imprinting (like NUP98-NSD1), or by directing transcriptional machinery to developmentally inappropriate targets (as seen in NUP98-KDM5A fusions and chromatin reader fusions). The mutational, structural, transcriptional, and epigenomic signatures of these two major groups of NUP98 fusion partners are so starkly distinct that one cannot help but speculate that each group should be treated as a separate subtype of AML, where both common and rare partners are likely to respond to similar treatments, whether repurposed (disulfiram for chromatin reader fusions) or novel (CDK9 inhibitors for HOX fusions).
Overall, NUP98 fusions constitute a highly refractory class of AML, which justifies reclassification of NUP98 fusions, regardless of fusion partner, as a high-risk subtype in future trials. Further research may focus on NUP98 fusion cases with aberrations of chr13, typically co-occurring with NUP98 exon 13 breakpoints and a distinctive immunophenotype, whose outcomes are relatively favorable given standard of care induction and/or transplantation. The balance of NUP98 fusions, with or without characteristic co-occurring mutations, remains an urgent, unmet therapeutic need. The immunophenotype, transcriptome, and epigenome of HOX-activating (versus chromatin-reader) fusion partners may provide important leads towards more effective therapies, while their signatures may permit rapid discontinuation of ineffective therapies in this high-risk group of patients.
Supplementary Material
Acknowledgments
The authors wish to gratefully acknowledge the important contributions of the late Dr. Stephen H. Petersdorf to SWOG and to the study S0106.
Funding Statement
Funding: This work was supported by the following NIH/NCI/NCTN grant awards: RO1CA190661, R01CA160872, R01AI171984, U10CA180888, U10CA180819, and U24CA196175, U10CA180886, U10CA180899, St. Baldricks Foundation, the Rally Foundation, and the Michelle Lunn Hope Foundation. This content is solely the responsibility of the authors and does not necessarily represent the ofcial views of the National Institutes of Health.
References
- 1.Balgobind BV, Hollink IH, Arentsen-Peters ST, et al. Integrative analysis of type-I and type-II aberrations underscores the genetic heterogeneity of pediatric acute myeloid leukemia. Haematologica. 2011;96(10):1478-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zwaan CM, Kolb EA, Reinhardt D, et al. Collaborative efforts driving progress in pediatric acute myeloid leukemia. J Clin Oncol. 2015;33(27):2949-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Rooij JD, Zwaan CM, van den Heuvel-Eibrink M. Pediatric AML: from biology to clinical management. J Clin Med. 2015;4(1):127-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Creutzig U, Zimmermann M, Ritter J, et al. Treatment strategies and long-term results in paediatric patients treated in four consecutive AML-BFM trials. Leukemia. 2005;19(12):2030-2042. [DOI] [PubMed] [Google Scholar]
- 5.Michmerhuizen NL, Klco JM, Mullighan CG. Mechanistic insights and potential therapeutic targets for NUP98-rearranged hematologic malignancies. Blood. 2020;136(20):2275-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura T, Largaespada DA, Lee MP, et al. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet. 1996;12(2):154-158. [DOI] [PubMed] [Google Scholar]
- 7.Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood. 2011;118(24):6247-6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Struski S, Lagarde S, Bories P, et al. NUP98 is rearranged in 3.8% of pediatric AML forming a clinical and molecular homogenous group with a poor prognosis. Leukemia. 2017;31(3):565-572. [DOI] [PubMed] [Google Scholar]
- 9.Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood. 2011;118(13):3645-3656. [DOI] [PubMed] [Google Scholar]
- 10.de Rooij JD, Branstetter C, Ma J, et al. Pediatric non-Down syndrome acute megakaryoblastic leukemia is characterized by distinct genomic subsets with varying outcomes. Nat Genet. 2017;49(3):451-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaju RJ, Fidler C, Haas OA, et al. A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood. 2001;98(4):1264-1267. [DOI] [PubMed] [Google Scholar]
- 12.de Rooij JD, Hollink IH, Arentsen-Peters ST, et al. NUP98/JARID1A is a novel recurrent abnormality in pediatric acute megakaryoblastic leukemia with a distinct HOX gene expression pattern. Leukemia. 2013;27(12):2280-2288. [DOI] [PubMed] [Google Scholar]
- 13.Noort S, Wander P, Alonzo TA, et al. The clinical and biological characteristics of NUP98-KDM5A in pediatric acute myeloid leukemia. Haematologica. 2020;106(2):630-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lange BJ, Smith FO, Feusner J, et al. Outcomes in CCG-2961, a children's oncology group phase 3 trial for untreated pediatric acute myeloid leukemia: a report from the children's oncology group. Blood. 2008;111(3):1044-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper TM, Franklin J, Gerbing RB, et al. AAML03P1, a pilot study of the safety of gemtuzumab ozogamicin in combination with chemotherapy for newly diagnosed childhood acute myeloid leukemia: a report from the Children's Oncology Group. Cancer. 2012;118(3):761-769. [DOI] [PubMed] [Google Scholar]
- 16.Aplenc R, Meshinchi S, Sung L, et al. Bortezomib with standard chemotherapy for children with acute myeloid leukemia does not improve treatment outcomes: a report from the Children's Oncology Group. Haematologica. 2020;105(7):1879-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollard JA, Loken M, Gerbing RB, et al. CD33 expression and its association with gemtuzumab ozogamicin response: results from the randomized phase III Children's Oncology Group Trial AAML0531. J Clin Oncol. 2016;34(7):747-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562(7728):526-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ley TJ, Miller C, Ding L, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson JE, Kopecky KJ, Willman CL, et al. Outcome after induction chemotherapy for older patients with acute myeloid leukemia is not improved with mitoxantrone and etoposide compared to cytarabine and daunorubicin: a Southwest Oncology Group study. Blood. 2002;100(12):3869-3876. [DOI] [PubMed] [Google Scholar]
- 21.Petersdorf SH, Rankin C, Head DR, et al. Phase II evaluation of an intensified induction therapy with standard daunomycin and cytarabine followed by high dose cytarabine for adults with previously untreated acute myeloid leukemia: a Southwest Oncology Group study (SWOG-9500). Am J Hematol. 2007;82(12):1056-1062. [DOI] [PubMed] [Google Scholar]
- 22.Godwin JE, Kopecky KJ, Head DR, et al. A double-blind placebo-controlled trial of granulocyte colony-stimulating factor in elderly patients with previously untreated acute myeloid leukemia: a Southwest Oncology Group study (9031). Blood. 1998;91(10):3607-3615. [PubMed] [Google Scholar]
- 23.List AF, Kopecky KJ, Willman CL, et al. Benefit of cyclosporine modulation of drug resistance in patients with poor-risk acute myeloid leukemia: a Southwest Oncology Group study. Blood. 2001;98(12):3212-3220. [DOI] [PubMed] [Google Scholar]
- 24.Haas BJ, Dobin A, Li B, et al. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol. 2019;20(1):213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robertson G, Schein J, Chiu R, et al. De novo assembly and analysis of RNA-seq data. Nat Methods. 2010;7(11):909-912. [DOI] [PubMed] [Google Scholar]
- 26.Tian L, Li Y, Edmonson MN, et al. CICERO: a versatile method for detecting complex and diverse driver fusions using cancer RNA sequencing data. Genome Biol. 2020;21(1):126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson JT, Thorvaldsdóttir H, Wenger AM, Zehir A, Mesirov JP. Variant review with the integrative genomics viewer. Cancer Res. 2017;77(21):e31-e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson JT, Thorvaldsdóttir H, Turner D, Mesirov JP. igv.js: an embeddable JavaScript implementation of the Integrative Genomics Viewer (IGV). Bioinformatics. 2023;39(1):btac830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edmonson MN, Zhang J, Yan C, et al. Bambino: a variant detector and alignment viewer for next-generation sequencing data in the SAM/BAM format. Bioinformatics. 2011;27(6):865-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chisholm KM, Heerema-McKenney AE, Choi JK, et al. Acute erythroid leukemia is enriched in NUP98 fusions: a report from the Children's Oncology Group. Blood Adv. 2020;4(23):6000-6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eidenschink Brodersen L, Alonzo TA, Menssen AJ, et al. A recurrent immunophenotype at diagnosis independently identifies high-risk pediatric acute myeloid leukemia: a report from Children's Oncology Group. Leukemia. 2016;30(10):2077-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ostronoff F, Othus M, Gerbing RB, et al. NUP98/NSD1 and FLT3/ITD coexpression is more prevalent in younger AML patients and leads to induction failure: a COG and SWOG report. Blood. 2014;124(15):2400-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Traag VA, Waltman L, van Eck NJ. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep. 2019;9(1):5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiba N, Ohki K, Kobayashi T, et al. High PRDM16 expression identifies a prognostic subgroup of pediatric acute myeloid leukaemia correlated to FLT3-ITD, KMT2A-PTD, and NUP98-NSD1: the results of the Japanese Paediatric Leukaemia/Lymphoma Study Group AML-05 trial. Br J Haematol. 2016;172(4):581-591. [DOI] [PubMed] [Google Scholar]
- 37.Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dard A, Reboulet J, Jia Y, et al. Human HOX proteins use diverse and context-dependent motifs to interact with TALE class cofactors. Cell Rep. 2018;22(11):3058-3071. [DOI] [PubMed] [Google Scholar]
- 39.Thirant C, Ignacimouttou C, Lopez CK, et al. ETO2-GLIS2 hijacks transcriptional complexes to drive cellular identity and self-renewal in pediatric acute megakaryoblastic leukemia. Cancer Cell. 2017;31(3):452-465. [DOI] [PubMed] [Google Scholar]
- 40.Huang BJ, Smith JL, Wang YC, et al. CBFB-MYH11 fusion transcripts distinguish acute myeloid leukemias with distinct molecular landscapes and outcomes. Blood Adv. 2021;5(23):4963-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanochko D, Halabelian L, Henderson E, et al. Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex. Nucleic Acids Res. 2019;47(3):1225-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heikamp EB, Henrich JA, Perner F, et al. The Menin-MLL1 interaction is a molecular dependency in NUP98-rearranged AML. Blood. 2022;139(6):294-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rasouli M, Szoltysek K, Cameron R, et al. NUP98/NSD1-positive AML is addicted to functional Menin-MLL interaction. EHA Library. 06/09/21;325136;EP382. [Google Scholar]
- 44.Bolouri H Farrar JE Triche T, Jr.et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. 2018;24(1):103-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.