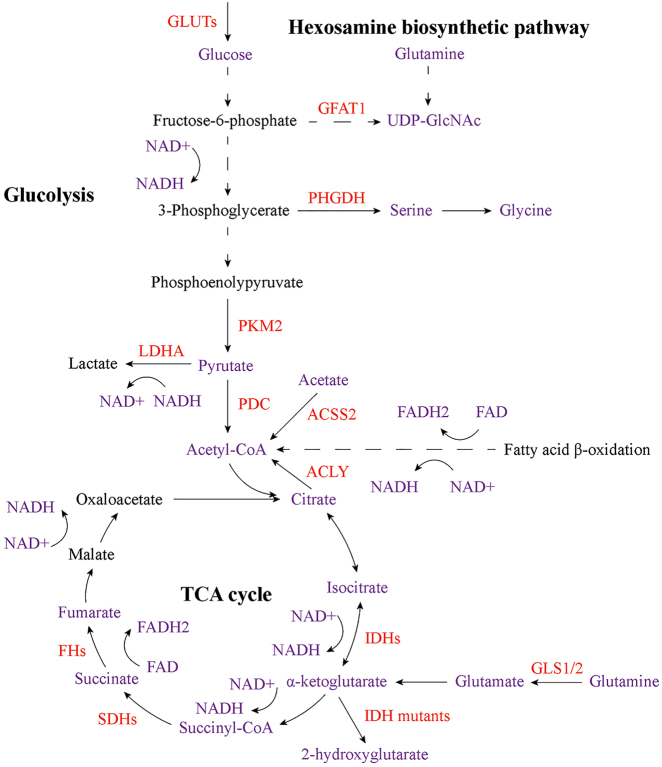
Figure 3:
Core cellular metabolic reprogramming that contributes to altered epigenetic modifications in cancer. Changes in metabolic proteins, which play important roles in glycolysis, TCA cycle and hexosamine biosynthetic pathway, affect the abundance of key metabolites, including acetyl-CoA, α-KG, succinate, fumarate, 2-hydroxyglutarate, UDP-GlcNAc, lactate and NAD+/NADH. These alterations have significant influences on DNA, RNA and histone methylation as well as histone acetylation, O-GlcNAcylation, phosphorylation and lactylation in cancer cells. Red indicates altered metabolic proteins, and purple indicates the key metabolites that affect epigenetic modifications in cancer. GLUTs, glucose transporters; GFAT1, glutamine fructose-6-phosphate amidotransferase 1; UDP-GlcNAc, uridine diphospho-N-acetylglucosamine; PHGDH, phosphoglycerate dehydrogenase; PKM2, pyruvate kinase 2; LDHA, lactate dehydrogenase A; PDC, pyruvate dehydrogenase complex; ACSS2, acyl-CoA synthetase short chain family member 2; ACLY, ATP-citrate lyase; IDHs, isocitrate dehydrogenases; SDHs, succinate dehydrogenases; FHs, fumarate hydratases; GLS1/2, glutaminase 1 and 2.