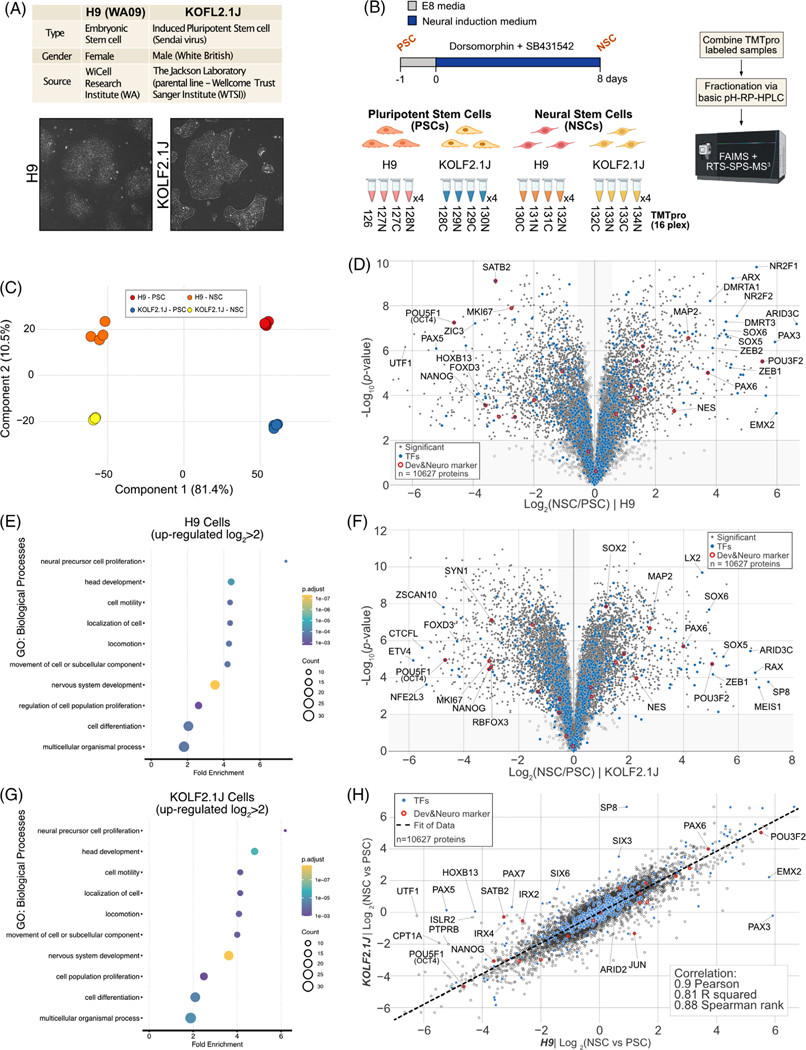
FIGURE 1.
Quantitative comparison of H9 and KOLF2.1 proteome remodeling upon Neural Stem Cell differentiation. (A) The information of H9 and KOLF2.1J reference cell lines (upper). Bright-field microscope images of H9 and KOLF2.1J cell lines (lower). (B) Workflow for neural stem cell differentiation from pluripotent stem cell and TMTpro based proteomics. 16plex proteomics was performed with biological quadruplicates per condition. (C) Principal component analysis (PCA) of proteomics data obtained from 16plex TMTpro based analysis. The inset indicates color coding for individual cells (Pluripotent Stem cell (PSC), Neural Stem Cell (NSC). (D) Volcano plot shows the H9 cell line -log10-transformed p-value versus the log2-transformed ratio of Neural Stem cells/ Pluripotent Stem cells for quantification. p-values were calculated by two-sided Welch’s t-test (adjusted with 1% false discovery rate (FDR) for multiple comparisons); for parameters, individual p-values and q-values, see Table S1. Individual proteins are shown in gray open circles, significantly changed protein: filled gray circle, transcription factors (TFs): filled blue circles, and developmental & neuronal markers: red open circles; 10627 proteins were quantified). (E) The enriched gene ontology (GO) terms for proteins whose abundance is significantly increased in H9 NSC compared to H9 PSC. (F) Volcano plot shows the KOLF2.1J cell line -log10-transformed p-value versus the log2-transformed ratio of Neural Stem cells/ Pluripotent Stem cells for quantification. p-values were calculated by two-sided Welch’s t-test (adjusted with 1% FDR for multiple comparisons); for parameters, individual p-values and q-values, see Table S1. Individual proteins are shown in gray open circles, significantly changed protein: filled gray circle, transcription factors (TFs): filled blue circles, and developmental and neuronal markers: red open circles, 10627 proteins were quantified). (G) The enriched gene ontology (GO) terms for proteins whose abundance is significantly increased in KOLF2.1J NSC compared to KOLF2.1J PSC. (H) Proteome level correlation plot of log2(NSC/PSC) from KOLF2.1J cell line (y axis) or H9 cell line (x axis) upon differentiation. The calculated correlation factors (Pearson, R squared, and Spearman rank) are indicated