Fig. 9. In silico recommendation for obtaining a 4-layered structure.
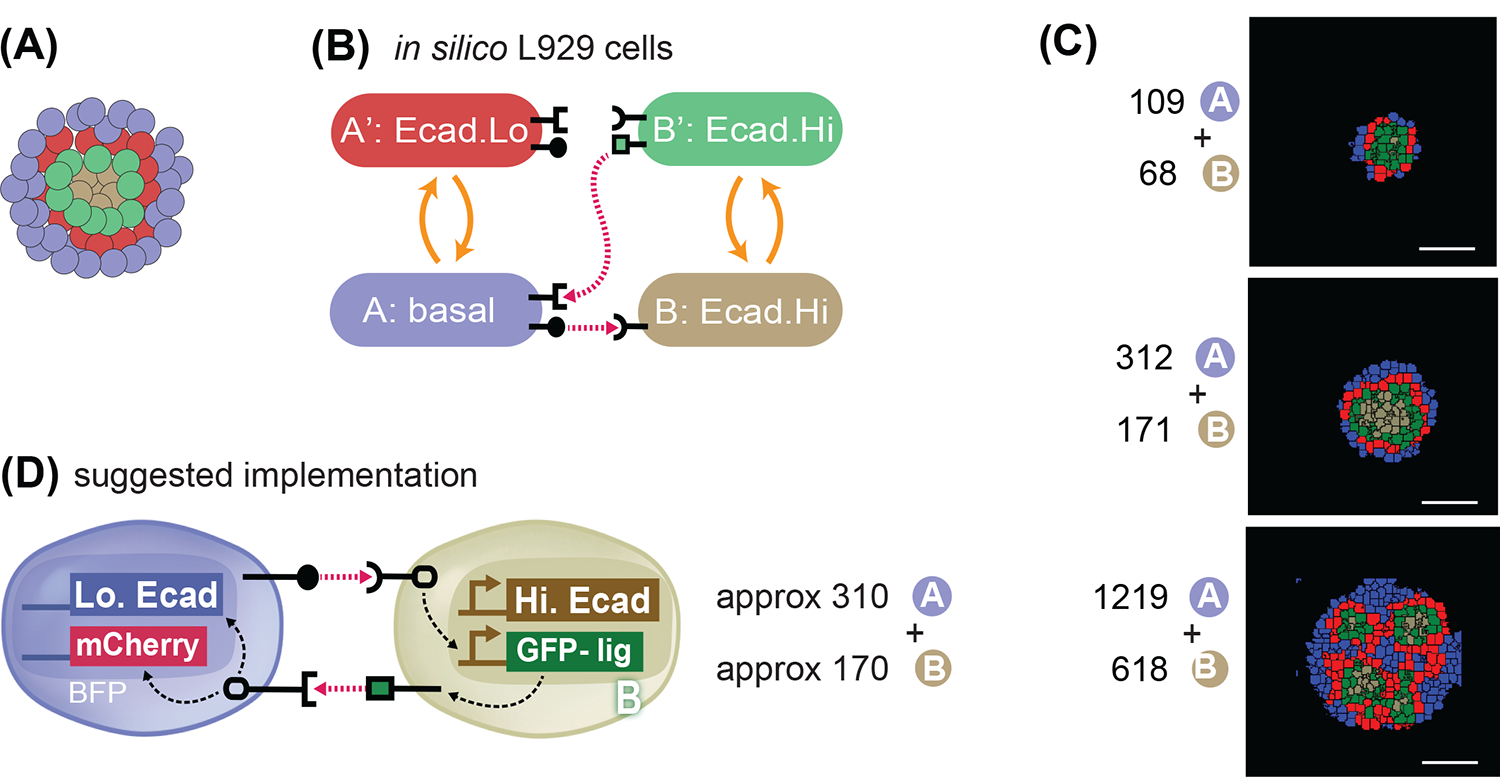
(A) Goal target pattern: 4 layers with gray-green-red-blue cells centrally symmetric. (B) State-machine diagram of the network, with the adhesion components. B cell has adhesion with Ecad.Hi parameter, and rounded receptor; when it is activated by a neighboring rounded receptor it activates to a B’ state that is adhesive with the same Ecad.Hi parameter, and produces a square ligand; A cell, blue, has basal adhesion, expresses rounded ligand, and a square receptor; the activation of the square receptor activates A cell to A’, which is red and has adhesion of Ecad.Lo parameter. (C) Representative results of resulting structures at t=24h of simulated time of cells with the genotype as in (9A) and with the ratio and numbers as indicated. All structures generate inactive A cells, activated A’ cells, inactive B cells and activated B’ cells in different ratios and in different geometrical arrangements. See Supplem. Fig. S6 for additional replicates over 10 runs of the simulation with the same parameters and initial conditions. (D) Recommended in vitro implementation. Scale bar is 17.5pixels in silico, equivalent to approx. 100um in vitro.
