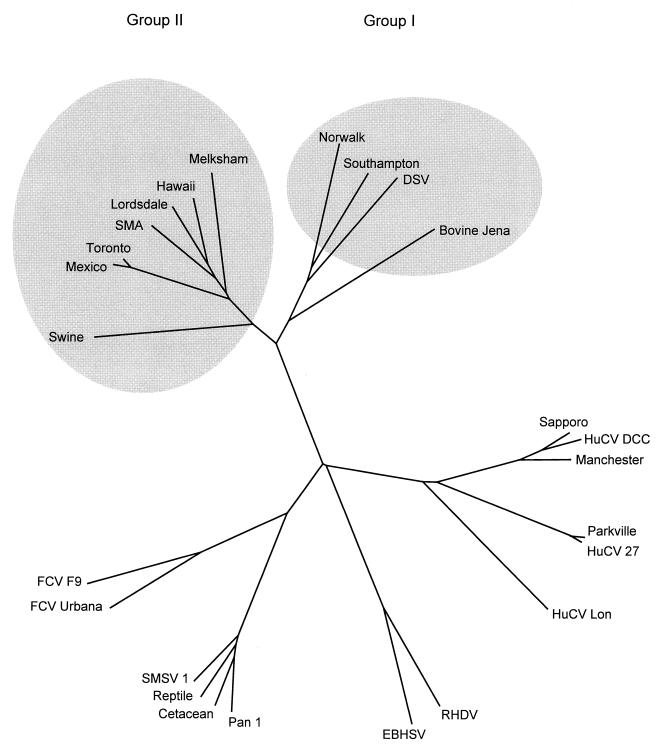
FIG. 4.
Unrooted phylogenetic tree constructed for a region of the 3D RNA-dependent RNA polymerase gene showing the relationship of JV to other caliciviruses. Shaded ellipses have been added to highlight the distinction between the group 1 and group 2 NLVs. Accession numbers (in parentheses) for caliciviruses are as follows; swine (AB009412); Mexico (U22498); Toronto (U02030); SMA (L23831); Lordsdale (X86557); Hawaii (U07611); Melksham (X81879); Norwalk (M87661); Southampton (L07418); Desert Shield virus (DSV) (U04469); bovine Jena (AJ011099); Sapporo (S77903); human calicivirus (HuCV) DCC (U67856); Manchester (X86559); Parkville (U73124); HuCV 27 (U67859); HuCV Lon (U67858); rabbit hemorrhagic disease virus (RHDV) (M67473); European brown hare syndrome virus (EBHSV) (Z69620); Pan 1 (U52086); cetacean (U52091); reptile (U52092); San Miguel sea lion virus (SMSV 1) (M87481); feline calicivirus (FCV) Urbana (L40021); FCV F9 (M86379).