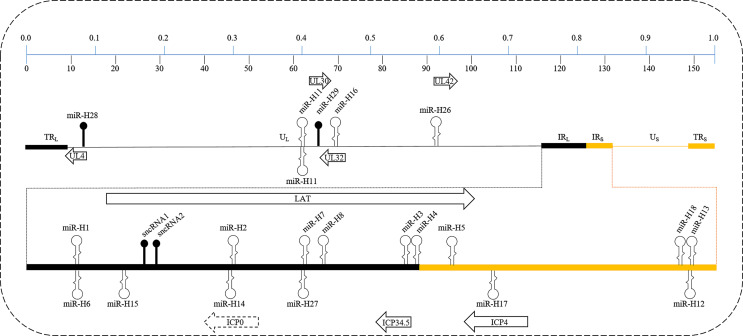
Fig. 1.
Schematic of the HSV-1 strain 17 genome (NC_001806) expanded to display the details of internal repeat unique sequences of long (IRL) and internal repeat unique sequences of short (IRS) loci. The upper and lower panels indicate the percentage of the coordinated genome and exact kilobase nucleotides, respectively. The locations of pre-miRNAs in the viral genome are shown as hairpins with relative sizes; pre-miRNAs shown above the line are located in the plus strand, which are transcribed from left to right (in the same direction as LAT), whereas those below the line are located in the minus strand, which are transcribed from right to left (in the opposite direction as LAT). All pre-miRNAs have stem-loop structures except for miR-H28, miR-H29, sncRNA1, and sncRAN2, which are indicated by the icons of black solid balls. Additionally, pre-miR-H11 comprises a 130 bp perfectly inverted repeat sequence. TRL, terminal repeat sequence of long; TRS, terminal repeat sequence of short; UL, unique long; US, unique short.