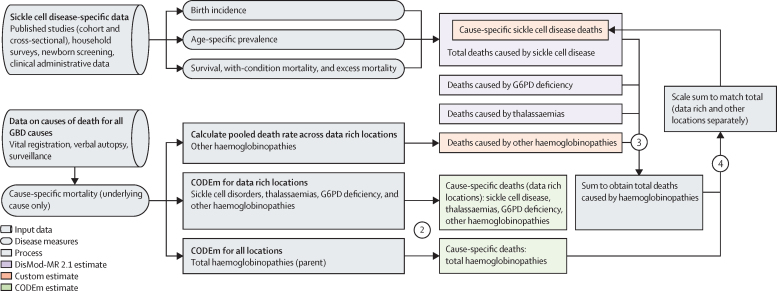
Figure 1.
Flowchart showing prevalence, cause-specific mortality, and total sickle cell disease mortality estimation process
Overview of the estimation process for sickle cell disorders for both cause-specific mortality on the basis of the International Classification of Diseases definition of one cause per death and total sickle cell disease mortality. Shapes differentiate input data, disease measures, processes, and estimates, whereas colours distinguish different modelling tools. The four steps are highlighted: (1) sickle cell disease-specific data on disease epidemiology were inputted into DisMod-MR 2.1 models to generate internally consistent incidence, prevalence, and overall mortality; (2) causes of death data processed across GBD were run through a series of CODEm to generate mortality estimates for total haemoglobinopathies (parent GBD cause) for all locations and for data rich locations only, for each of the child causes, including sickle cell disease, thalassaemias, G6PD deficiency, and other haemoglobinopathies; (3) the mortality results from each of the four subcauses were scaled to 100%; and (4) these mortality results were used to divide total haemoglobinopathies CODEm results into cause-specific mortality results for sickle cell disease and other subcauses. CODEm=Causes of Death Ensemble model. G6PD=glucose-6-phosphate dehydrogenase.