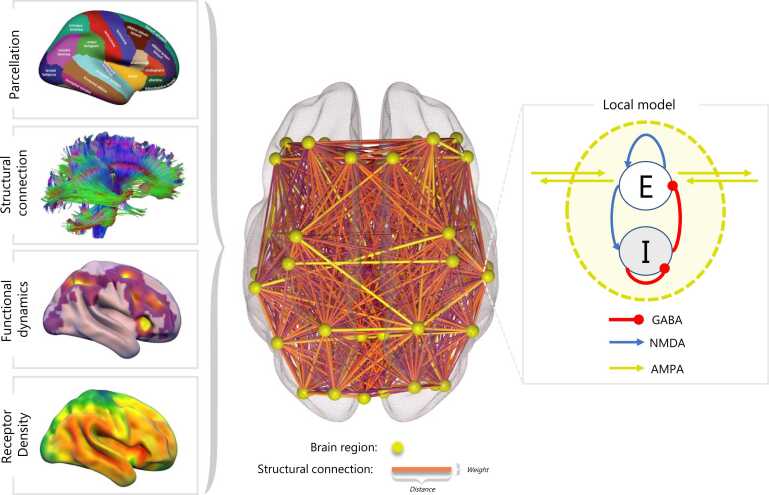
Fig. 3.
An overview of the necessary multimodal constituents for a whole brain computation model. A parcellation defines a series of nodes (brain regions) which are connected based upon structural connectivity from diffusion tensor imaging (DTI). Each node is modelled through biophysically plausible excitatory (E; NMDA) and inhibitory (I; GABA) dynamics as shown in the local model on the right. In addition, molecular density information from PET can be incorporated into the model to shape specific aspects of the model such as neuronal gain.