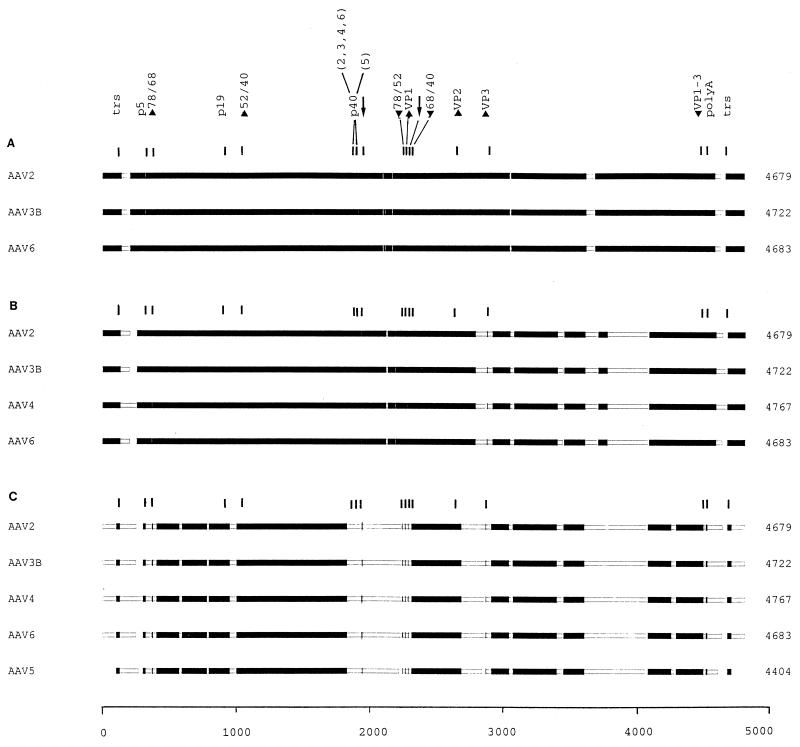
FIG. 3.
Block alignment of AAV genomic sequences. The complete sequences of AAV2, AAV3B, AAV4, and AAV6 and the partial sequence of AAV5 were aligned with the MACAW program. Segment pair overlap with a minimum segment pair score of 60 was used as a search method to define nucleotide blocks of local similarity. The aligned blocks were linked into the alignment (solid bars). To determine the start and end positions for the alignment of AAV5, the trs (50) were selected by sequence analogy. The scale of alignment is 5,000 nt. Nucleotide positions on the individual genomes are in relation to this scale. The panels show the alignment of AAV2, AAV3B, and AAV6 (A); the alignment of AAV2, AAV3B, AAV4, and AAV6 (B); and the alignment of AAV2, AAV3B, AAV4, AAV6, and AAV5 (C). The positions of the conserved genetic elements are marked, including the p5, p19, and p40 promoters (position of TATA boxes); the intron splice sites (GT/AG [arrows]); the polyadenylation signal (polyA); the translation start (up arrowheads) and stop (down arrowheads) sites for the Rep (Rep 78, 68, 52, and 40) and capsid (VP1 to VP3) proteins; and the terminal resolution site and its inverted counterpart (both marked trs). Note that all of the elements in panel A were within blocks defined by the program and that in panel B only the start of VP3 was excluded, while in panel C several were no longer in regions that met the criteria required for block selection [p5, p40, start Rep 78 and 68, stop Rep 78 and 52, start VP1 and VP3, the splice signals, and the poly(A) site]. Such positions were marked manually. Note that the p40 sites in panel C are located in stretches of high dissimilarity and were not aligned.