Summary
In recent years, the amount of data generated by imaging techniques has grown rapidly, along with increasing computational power and the development of deep learning algorithms. To address the need for powerful automated image analysis tools for a broad range of applications in the biomedical sciences, the Microscopic Image Analyzer (MIA) was developed. MIA combines a graphical user interface that obviates the need for programming skills with state-of-the-art deep-learning algorithms for segmentation, object detection, and classification. It runs as a standalone, platform-independent application and uses open data formats, which are compatible with commonly used open-source software packages. The software provides a unified interface for easy image labeling, model training, and inference. Furthermore, the software was evaluated in a public competition and performed among the top three for all tested datasets.
Keywords: image analysis, deep learning, microscopy, segmentation, classification, object detection, tracking
Graphical abstract
Highlights
-
•
MIA is an open-source software solution for image analysis
-
•
Graphical user interface for easy access to powerful algorithms
-
•
Image labeling, training of deep neural networks, and prediction of unseen data
-
•
Applications for classification, semantic segmentation, object detection, and tracking
Motivation
In recent years, advances in computer vision and automated image analysis have led to major scientific breakthroughs. However, widespread use for microscopic image analysis has been limited by the technical expertise required for their use. MIA software has been developed in an effort to provide access to state-of-the-art image analysis for all researchers.
Körber presents a software tool that lowers the entry level for deep-learning-based image analysis, making it accessible to researchers regardless of their scientific background. The software enables labeling, model training, and prediction using image data.
Introduction
Deep neural networks are the state of the art for most computer vision tasks and often outperform classical image analysis techniques.1 In recent years, deep learning has slowly evolved from the field of computer science into applications in microscopy, achieving impressive results in areas such as cancer detection,2,3 super-resolution,4 denoising,5,6 and label-free staining.7,8 With the massive growth of image data generated by microscopy and high-throughput imaging, the demand for automated image analysis solutions has increased drastically. So far, the implementation and training of deep learning algorithms for image analysis has required advanced coding skills, mathematical expertise, and computational resources, limiting its broad application in the life science community. Various solutions have been deployed to reduce these hurdles and provide an easily accessible interface for deep learning, such as DeepCell Kiosk,9 ImJoy,10 ZeroCostDL4Mic,11 the U-Net plugin,12 ilastik,13 CellProfiler,14 3DeeCellTracker,15 DeepImageJ,16 CellPose,17 Stardist,18 CDeep3M,19 nucleAIzer,20 and more. DeepCell Kiosk,9 ImJoy,10 and CDeep3M19 provide web applications that can be easily accessed using a browser and often provide pre-trained solutions for specific tasks. Web applications offer a low barrier to entry and easy exchange of models; however, they rely on third-party hardware, and customization remains limited. ZeroCostDL4Mic11 and 3DeeCellTracker15 offer implemented algorithms via interactive notebooks that can be run on individual datasets. The interactive interface in combination with direct access to the code and documentation allow researchers to adapt an existing pipeline to their needs. On the other hand, the adaptation requires actual writing of source code and might be cumbersome for inexperienced users. Additionally, notebooks lack an interactive interface when it comes to image labeling or browsing images. U-Net,12 DeepImageJ,16 CellPose,17 nucleAIzer,20 and Stardist18 are available on the command line or as plugins. Each plugin offers a solution for a specific task, such as segmentation of cells with high accuracy. The extension to broader applications remains difficult, and the interoperability of the plugin depends on the software in which it was deployed. CellProfiler,14 ilastik,13 and ImageJ21 are complete software packages addressing a broad range of applications with a plethora of tools and out-of-the-box algorithms for image analysis. Regardless of the undoubtedly great contribution these programs have made to the imaging community, they were not designed primarily for deep learning applications. Although each of the aforementioned tools and applications provides great value for certain aspects of the deep learning procedure, such as training, cloud computation, and pre-trained model integration, a comprehensive, unified, and user-friendly tool that integrates these features into a single versatile software is lacking. Thus, my group developed MIA, the Microscopic Image Analyzer, as standalone software with a graphical interface integrating image labeling, model training, and inference that can be operated platform independently (Figure 1). MIA was designed with a particular focus on usability and simplicity. Although model training time is often considered critical, the most laborious part of the deep learning procedure is by far image labeling. Assisted labeling can significantly reduce the required hands-on working hours. Whereas comparable applications require expert knowledge or have very limited access to complex features, MIA offers them as selectable built-in options. Users can choose from a number of neural network architectures, with several training options, providing experienced users with the option to customize for a specific dataset while still achieving good results with default settings. The training computation can be performed by the graphical processing unit (GPU) or the central processing unit (CPU), depending on the available hardware. The training process and model performance are monitored during training, and the prediction of unseen data can be visually inspected and corrected if necessary. Finally, the results can be saved for individually detected objects, and labels can be exported for further analysis with other software.
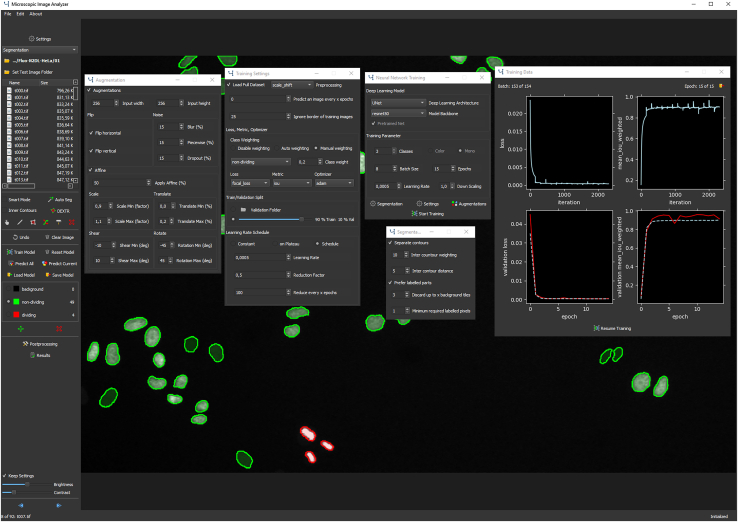
Figure 1.
The MIA user interface
The left panel of the user interface shows an explorer interface for file navigation, tools for image labeling and options for model training, prediction, saving, loading, post-processing, and setup of object classes and other options. The center panel shows the currently displayed image with labeled cells, overlaid with several open windows showing training options, augmentation settings, training progress, and others. The currently displayed image shows HeLa cells expressing H2b-GFP.22
Results
MIA features
The motivation for the development of MIA was to create a tool that gives any user access to powerful deep learning image analysis in order to address different types of scientific questions. However, to cover a wide range of applications and imaging modalities, training parameters and technological depth still need to be adjustable to achieve excellent results for each use case. As easy access to deep learning competes with freedom and performance, MIA lowers the entry level for deep learning at the expense of flexibility. See Table 1 for a comparison of MIA with other methods used in microscopic image analysis. The scope of MIA is the analysis of microscopic data with a user-friendly interface (see Tables S1 and S2 for the results of a usability test). To cover the most common tasks of image analysis, image classification, semantic segmentation, and object detection have been implemented, along with a tracking mode as post-processing of previously detected objects. To test and demonstrate the features of MIA, training datasets for classification, segmentation, and object detection were used from different imaging domains and applications. In the following sections, these datasets are used to showcase different applications of MIA. Each application was trained using the default settings of MIA, without any adjustments, to show that even non-experts can achieve convincing results.
Table 1.
Comparison of MIA with classical image analysis and deep learning
| Classical analysis | Deep learning | MIA | |
|---|---|---|---|
| Requirements | |||
| A priori domain knowledge | medium to high | low | low |
| Training data | none or few | medium to high | medium to high |
| Training/processing time | low | high | high |
| Coding skills/mathematical expertise | low to medium | high | none |
| Development effort | low to medium | high | low |
| Image labeling tools | not required | external | included |
| Capabilities | |||
| Improves with increasing data | no | yes | yes |
| Generalization | low | high | high |
| Performance | low to medium | high | high |
| Flexibility | medium | high | medium |
| Application range | medium | high | medium |
| Revision of predictions | external | external | included |
Image classification
Image classification is the categorization of each image in a predefined class. For example, it can be used to identify different cell types or different phenotypes following drug treatment. As a complete image is assigned to only a single class label, information on where in the image a particular object is located is not considered, yet the labeling process is much faster compared with labeling of individual pixels. For classification with MIA, the user can choose from a variety of implemented network architectures, ranging from fast networks such as mobilenet23 to larger and slower models like NASNet.24 To showcase the capabilities of MIA for classification, a neural network was trained on the PCam dataset (Figure 2B).25,26 The dataset comprises patches (96 × 96 px) derived from histopathological scans of lymph node sections that are classified as metastatic or normal tissue. MIA was used to train a baseline network on the training images of the dataset and was evaluated on images not included during training. The model achieved accuracy of 86% ± 2% to correctly distinguish healthy from malignant tissue.
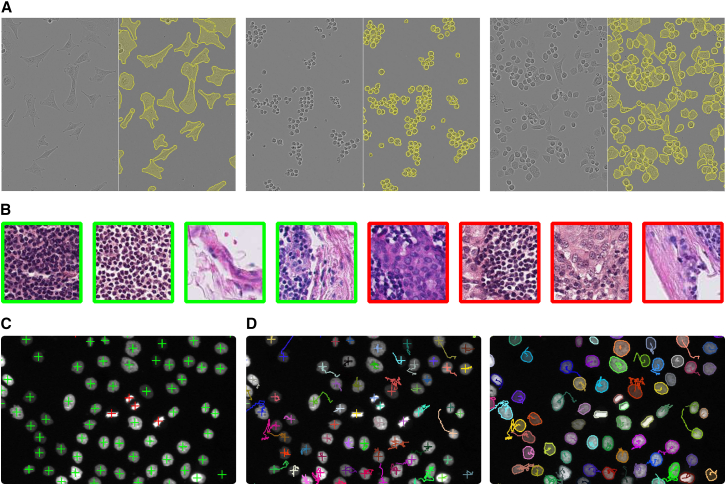
Figure 2.
Example applications using MIA
(A–D) Neural networks were trained with MIA default settings for segmentation (A), classification (B), object detection (C), and tracking (D) using different datasets.
(A) A neural network was trained to segment individual cells from light microscopy using the LIVEcell dataset.27 The model achieved an intersection over union (IoU) of 0.8 ± 0.01 on the test data. Example images from the test data are shown in pairs, with the original image on the left and the same image with the model predictions overlaid in yellow on the right.
(B) A neural network was trained to classify lymph node tissue in normal (green) or containing metastatic tissue (red).25,26 The model achieved accuracy of 86% ± 2% to correctly classify metastatic tissue in the test data.
(C) A neural network trained for object detection achieved a mean absolute error (MAE) of 0.5 ± 0.01 on the identification of dividing (red) and non-dividing (green) cells expressing H2b-GFP.22
(D) Results from object detection or segmentation can be used to perform tracking of individual objects over consecutive time points. Each object is shown in an individual color and traces refer to positions along previous time points. Results are reported as the mean ± SD of 3 training runs.
Semantic segmentation
Semantic segmentation is the process of assigning a predefined class to every pixel of an image. It allows foreground and background as well as different cell or tissue types to be distinguished. The number of classes need to be defined prior to training, and all objects need to be labeled with their corresponding class with the unlabeled area assigned to background. MIA provides several labeling tools for semantic segmentation, such as polygon tool, freeform tool, slicing tool, an extension tool, and an erase tool. However, to label objects one by one can be tedious for complex structures and large numbers of objects. Hence, automated or semi-automated labeling tools have been implemented on the basis of Grabcut,28 holistically nested edge detection,29 and Deep Extreme Cut,30 that may reduce labeling time drastically.30 Several neural network architectures for semantic segmentation, including the popular U-Net31 and DeepLab,32 can be chosen building on a variety of selectable model backbones, from computational less expensive models such as mobilenetv223 to more complex models such as EfficientNet.33 To avoid fusion of neighboring objects, a common problem in the segmentation of dense microscopic images, MIA supports increasing the weighting of pixels in close proximity to adjacent objects, as shown by Ronneberger et al.31; in that way, the network learns to separate neighboring objects during training. To illustrate the applicability of MIA for semantic segmentation, a U-Net31 was trained to segment different cell types from phase-contrast microscopy.27 The training images show cells in culture and their corresponding pixel-precise outlines as labels (Figure 2A). The trained model achieved a mean intersection over union of 0.8 ± 0.01 for unseen data to correctly classify pixels of different cell types. The shape, size, and position of each cell can be extracted from the predicted image data.
Object detection
Object detection identifies all instances of all classes and their respective positions in the image. Its primary task is to count objects, for example, cells of different cell types in a tissue and their spatial distribution. In computer vision, object detection is often defined as the detection of each object instance along with its corresponding bounding box. However, in MIA, we designed object detection by the center position of the object omitting the bounding box, as in the life sciences the bounding box rarely offers relevant information. To measure the size of detected objects, it is recommended to use semantic segmentation instead. For object detection in MIA, the same model architectures can be chosen as for semantic segmentation, but a linear layer is used in the final layer of the network instead of a softmax or sigmoid layer, enabling the regression of peak values at image spots where objects are located. To exemplify the use of object detection, MIA was used to train a model for the detection of dividing and non-dividing HeLa cells expressing H2b-GFP.22 The dataset comprises training images of cells and corresponding labels with the position information of dividing and non-dividing cells (Figure 2C). The trained model achieved a mean absolute error of 0.5 ± 0.01 on unseen images to correctly identify localizations of target cells. Exported results can be used for further analysis (e.g., to describe cell division events as a function of cell density).
Object tracking
Object tracking is the identification of the same object at consecutive time steps. One application of object tracking is to determine the trajectories of living cells in tissue or during development. Tracking is implemented as a post-processing step and can be used in combination with object detection or semantic segmentation. The center of each object is calculated and compared with the centers in the subsequent frame by minimizing the total distance between the objects in the two frames on the basis of the Hungarian algorithm.34 An object that is not detected in several consecutive frames is considered lost. In addition to automatic tracking, MIA makes it possible to reassign detected instances to different objects to correct for potential errors. Furthermore, MIA supports tracking of different object types in parallel. To illustrate object tracking with MIA, the results for dividing and non-dividing HeLa cells from semantic segmentation and object detection were used to calculate the position of each object in a time sequence (Figure 2D). On the basis of the detected objects in the image sequence, the detections of the same object in sequential images were combined to obtain the position information of that object along the entire sequence. As a result, motion profiles of unique objects with respect to the image sequence could be identified. The results may be used for further analysis, to calculate, for example, the cell movement in relation to division events.
Recommended MIA workflow
MIA supports input data from all common public image types and formats including 16-bit depth images and z stacks. The images can be labeled according to the respective target application, i.e. classification, semantic segmentation, or object detection. The initial decision which objects to label and which classes of objects to choose have a significant impact on the final result and depend on the specific scientific question. Once images are labeled, the training process can be started by selecting a model architecture. All models can be trained from scratch or, alternatively, using pre-trained weights, which were derived from training on a large dataset like imagenet.35 This so-called transfer learning allows faster convergence for smaller datasets. Another way to increase the amount of training data is data augmentation (i.e., modifying the images of the training set by image operations such as flipping, translation, and scaling). MIA provides a number of possible augmentations that are automatically applied to the input images during training. Hyperparameters, such as batch size or a specific learning rate schedule, can be defined via the user interface. For model optimization, the most commonly used loss functions (e.g., cross-entropy) and optimizers (e.g., Adam36) can be selected depending on the particular application. To monitor the model performance, it is common practice to split the training set into a training and a validation set, which can be done in MIA either randomly or by specification of a data subset. Real-world datasets are usually not balanced well, meaning that the frequency of different objects can differ strongly within the dataset. To circumvent this class imbalance, several options are implemented in MIA such as automatic class weighting, removal of less labeled image parts during training or specific loss functions such as focal loss.37 Once the model training is finished, the trained model can be applied to predict unseen images. The inference is always specified by the selected application (e.g., a model trained for semantic segmentation yields a model that can be used to segment images that are similar to the images present in the training set). All predicted images can be visually inspected and corrected if objects were incorrectly classified. We propose the following workflow to create a neural network performing well on a real-world dataset (Figure 3):
-
(1)
Label a small set of images.
-
(2)
Train a model.
-
(3)
Repeat until the model performs sufficiently well on unseen data:
-
(a)
Predict unlabeled images and evaluate model performance.
-
(b)
(i) Correct the predicted image labels, (ii) label additional images, or (iii) skip this step.
-
(c)
Update training dataset and retrain the model.
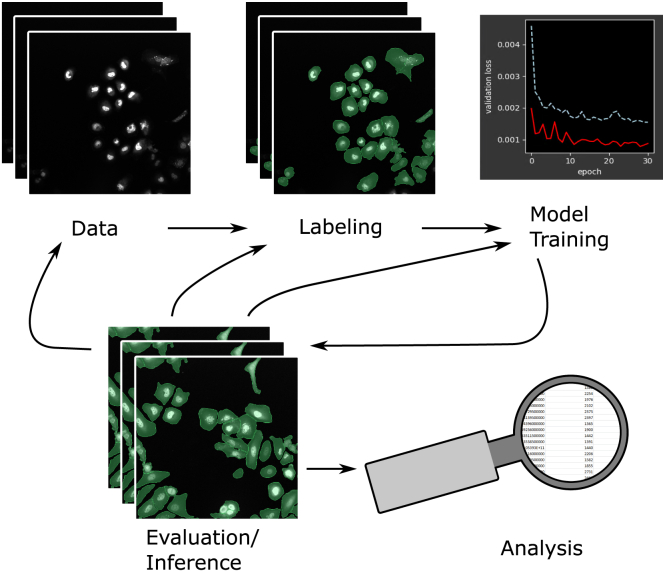
Figure 3.
Proposed workflow for efficient neural network training
Start with image labeling, train a model, and evaluate its performance. From there, label new data, correct predicted labels, or train a new model. Repeat this process until a sufficiently performing model is trained. Finally, the model can be used for prediction and analysis. Images show human hepatocarcinoma-derived cells expressing YFP-TIA-1.38
Note that training can be continued with conserved weights from previous training or started from scratch.
A significant advantage of this iterative approach with a human in the loop is that only rare cases need to be corrected, whereas correctly predicted labels can be reused without further work. This highly reduces the redundancy of the data and a large diversity of the dataset is granted. Thus, the labeling is focused on data that need supervision, whereas other parts for which the model is already performing well can be ignored. All trained models can be saved, reloaded, and further trained. Stored models always create identical predictions for the same input, so they can be used for archiving and quality control. Finally, detected objects can be analyzed on the basis of their size, shape, position, or signal intensity, and they can be exported for further analysis.
Cell segmentation benchmark
In order to evaluate the performance of MIA, the software was tested on datasets that are part of the Cell Tracking Challenge, a public competition for image analysis algorithms.39 The performance of MIA was compared with the state-of-the-art results of the cell segmentation benchmark (CSB). The challenge offers different datasets derived from two-dimensional (2D) or three-dimensional (3D) time-series experiments or simulations. Each dataset consists of a training set and a test set. The training set contains images and corresponding labels for segmentation, whereby the correctness and completeness of the labels varies for each dataset. The test set comprises only images, while the corresponding labels remain with the challenge organizers and are used for evaluation. The evaluation is based on the mean of an object detection score and an object-based intersection over union; see the Cell Tracking Challenge website for details.39 MIA was evaluated using three datasets each showing a different use case and relation to a real-world application. The first two datasets used for analysis with MIA contained complete but partly incorrect label information. The first dataset (Fluo-N2DH-SIM+)40 consists of simulated HL60 cells stained with Hoechst. Because of the nature of a simulated dataset, the image labels are exact and complete for the images of the training set. The second dataset (Fluo-N2DL-HeLa)22 consists of images of HeLa cells stably expressing H2b-GFP. The corresponding labels are computer generated from previous challenge submissions. In that way, all training images have corresponding labels, but labels may be incorrect for difficult cases. In order to enhance the quality of these labels, MIA was used to quickly screen through the data, and all labels that were obviously incorrect (i.e., missing a part of a target cell) were corrected by hand. For these two datasets, a DeepLabv3+ model architecture32 with a Xception backbone41 was chosen and trained on the training set. The trained models were used to evaluate their performance on the test set and achieved second and third rank of all submissions without bells and whistles (Table 2). However, real-world images usually have no inherently labeled objects. To further evaluate MIA in a more realistic scenario, the Fluo-C2DL-Huh7 dataset of human hepatocarcinoma-derived cells expressing the fusion protein YFP-TIA-1 was used.38 This training data consists of 58 images, of which only 13 are labeled. Furthermore, the difficulty of correctly segmenting the cells is higher compared with the other datasets because the cells are very dense, and their boundaries are visually indistinct (example images can be seen in Figure 3). To tackle this task, the workflow from the previous section was applied. A small model was trained on the existing 13 labeled images intentionally overfitting to the training data. This model was then used to predict the labels for the remaining unlabeled images of the training set. These predicted labels were corrected by hand utilizing MIA to create a complete training set. In addition, the task was treated as a three-class problem defining background, target cells, and the outer border of cells (three pixels wide) as individual object classes. Adjacent cells can be separated more easily when borders of cells are treated as an additional class since the network needs to explicitly identify the border region of cells. Finally, a U-Net31 with an Inception-v442 backbone was trained on the training data. With this approach, MIA achieved the new state of the art for this dataset (Table 2).
Table 2.
Top three results of the cell segmentation benchmark (OPCSB)39 for three datasets
| Rank | Fluo-N2DL-HeLa | FLUO-N2DH-SIM+ | Fluo-C2DL-Huh7 |
|---|---|---|---|
| 1 | 0.957 | 0.905 | 0.885∗ |
| 2 | 0.957∗ | 0.899 | 0.879 |
| 3 | 0.954 | 0.898∗ | 0.854 |
The top three results of the CSB are shown, with MIA results indicated by asterisks. See http://celltrackingchallenge.net/participants/BFR-GE/.
Discussion
With the development of MIA, we aim to simplify the use of deep learning for image analysis, with a particular focus on the diverse field of microscopy. MIA enables users without any programming experience to perform state-of-the-art image analysis on the basis of deep learning for a wide range of applications providing all necessary tools in one single application. Furthermore, the access to hyperparameters and training options can improve the understanding of deep learning methods and strengthen their acceptance. As MIA runs on local hardware, it is independent of any cloud service and can be scaled on the basis of resources and requirements. We found that most existing applications focus on a specific part of the deep learning sequence, such as training, inference, or a specific solitary task. On the other hand, a single application comprising all processing steps minimizes errors in data transfer and facilitates a seamless and optimized data flow. With MIA, we attempted to integrate the complete process, proposing a workflow with a human in the loop that is fast and flexible to create powerful models for most real-world applications. Default parameters in MIA depict the most commonly used options, thus inexperienced users are able to successfully train a model on any dataset, while users experienced in deep learning have all the options to customize the analysis to achieve state-of-the-art results. One major focus of MIA is image labeling, which is often not integrated in deep learning solutions but at the same time often is the most time-consuming part for the user in practice. MIA supplies automated solutions for image labeling that reduce hands-on time and the frustration of repetitive work. With the current version of MIA, it is possible to perform classification, object detection, segmentation as well as tracking, which covers a broad range of the microscopy image analysis portfolio. Future development could include the implementation of instance segmentation, 3D models, or automated hyperparameter tuning. All predictions generated by MIA can be exported as image mask for subsequent use with other image analysis software such as ImageJ21 and CellProfiler,14 placing the focus on deep learning while leaving downstream analysis or a larger image processing pipeline to external software. Along with this, trained models can be saved and reused with other applications or custom code using the TensorFlow SavedModel format,43 making the models portable and compatible for model exchange in accordance with FAIR (findability, accessibility, interoperability, and reusability) data principles.44 In summary, we provide a versatile tool that allows an easy access to state-of-the-art deep-learning-based image analysis for a broad range of applications.
Limitations of the study
Like all deep learning approaches, the MIA software requires training data. In cases in which the image source is limited and only a few training images can be generated, using the MIA software may not produce satisfactory results. In addition, using low-quality data or inaccurate labels will reduce the performance of the trained model. Furthermore, image labeling is time consuming, and it should be noted that in cases that can be solved with little effort (e.g., separating a good fluorescence signal from background), a simpler solution such as thresholding might lead to a faster solution for that specific problem.
In summary, optimal performance of the MIA software requires a good database with high-quality labels and sufficient computation time.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| LIVECell dataset | Edlund et al.27 | https://sartorius-research.github.io/LIVECell/ |
| PCam dataset | Bejnordi et al.25; Veeling et al.26 | https://github.com/basveeling/pcam |
| Fluo-C2DL-Huh7 dataset | Ruggieri et al.38 | http://celltrackingchallenge.net/2d-datasets/ |
| Fluo-N2DL-HeLa dataset | Neumann et al.22 | http://celltrackingchallenge.net/2d-datasets/ |
| Fluo-N2DH-SIM+ dataset | Svoboda et al.40 | http://celltrackingchallenge.net/2d-datasets/ |
| Software and algorithms | ||
| MIA (v 0.2.6) | This paper | https://doi.org/10.5281/zenodo.7970965 |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Nils Körber (koerbern@rki.de).
Materials availability
This study did not generate new unique reagents.
Method details
Environment
MIA is entirely written in Python using several publicly available open source solutions. The graphical user interface is written with PyQt (Riverbank Computing), the image processing and model computation are based mainly on Numpy,45 OpenCV,46 scikit-image,47 imgaug,48 TensorFlow43 and Keras.49 The model architectures are relying on open source repositories50,51,52,53 and custom implementations.
Requirements
MIA is cross-platform compatible and has been tested successfully on several systems running with different versions of Windows and Linux on workstations or servers. The software supports GPU acceleration of most CUDA54-compatible units. Even though training can be performed with a CPU, it is highly recommended to use a GPU to decrease training time significantly.
Datasets for the demonstration of MIA features
To showcase applications using MIA, different datasets for neural network training were used from different applications. For classification the PCam dataset for tumor detection was used.25,26 It comprises of 327.680 color images from histopathological lymph node scans, each 96 x 96px in size, classified in healthy and metastatic tissue. For training 262.144 training images were used and the model performance was evaluated on the 32.768 test images that do not overlap with the training images. The remaining validation images were not used during training or evaluation. The LIVEcell dataset, consisting of 3,727 training and 1,512 test images derived from phase-contrast microscopy of eight different cell lines, was used for segmentation.27 The resulting segmentations were separated using watershed algorithm55 on the predicted probabilities as implemented in the MIA software. The silver segmentation labels of HeLa cells expressing H2b-GFP22 provided by the Cell Tracking Challenge39 were used as starting point for label generation for object detection. MIA was used to introduce an additional third class to this binary labeled data. All dividing cells, i.e. cells that were split into two objects in the subsequent time point and two objects deriving from a single object from the previous time point, were reassigned to an extra class label. All remaining objects were kept unchanged, resulting in segmentation labels for dividing and non-dividing cells. Labels for object detection were generated from these segmentation labels by calculating the center of each contour using image moments. As validation data for object detection 15% of the training data were withhold from training. The neural network training for all datasets was performed using MIA’s default settings for the target application. All Predictions of images shown in Figure 2 belong to the test data.
Participation in the Cell Segmentation Benchmark
All steps used to participate in the Cell Tracking Challenge were done with the source code of MIA, except the conversion of the Cell Tracking Challenge labels to MIA compatible labels and vice versa. Since MIA counts touching objects as one, unlike the challenge, closely connected objects were separated by a zero-valued boundary between them when converted to MIA and expanded to the adjacent objects when converted back to the challenge labels to create touching objects. All models were trained with 512x512 image patches that are randomly sampled from the training images. The same augmentation strategy was used for all trainings using image flipping, rotation, up to 10% shearing, 90–110% scaling and a 15% probability of image blurring, piecewise affine image transformation or image dropout. The Adam optimizer36 with an initial learning rate of 0.0005, halved every 50 epochs, was used to optimize the cross entropy cost function. Fluo-N2DH-SIM+40: A DeepLabv3+32 with Xception41 backbone model was trained from scratch with a batch size of 16 for 500 epochs. The target cell class was weighted with 0.6 and background with 0.4 accordingly. Pixels between objects were weighted with a maximum weighting of ω0 = 10 and a border distance parameter of σ = 5 (as calculated in39). Fluo-N2DL-HeLa22: A DeepLabv3+32 with Xception41 backbone model with pre-trained weights was trained with a batch size of 16 for 300 epochs, using a class weighting of 0.9 to 0.1, a maximum inter-object pixel weighting of ω0 = 28 with a border distance parameter of σ = 5 (as calculated in39). Fluo-C2DL-Huh738: A U-Net39 with an Inception-v442 backbone with pre-trained weights was trained with a batch size of 8 for 500 epochs. The cell border class was weighted with 1, the center cell class with 0.1 and background with 0.05. All model predictions were done with test time augmentation, meaning that each input image was flipped and rotated by ±90° to generate a total of six input images. The model predictions of these transformed images were averaged to the final prediction. Post-processing to separate erroneously connected objects was either omitted or the detected objects were split based on watershed algorithm55 using the predicted class probabilities or based on morphological image operations.
Usability test
To test the usability of the MIA software, a usability test was conducted inspired by the System Usability Scale (SUS).56 A total of 13 participants (7 female, 6 male) from various backgrounds (medicine, biology, biosystems technology (2x), bioinformatics, mathematics, education, economics, chemistry, physics, computer science, engineering, unknown) who were unfamiliar with the software were given the task of labeling the contours of the nematode c.elegans on a small set of microscopic images for semantic segmentation. After labeling, participants were asked to train a deep neural network and use the trained network to predict images that were not used during training. Participants then answered a questionnaire related to the task (Table S1) and the MIA software in general (Table S2).
Quantification and statistical analysis
Neural network training was performed 3 times with identical settings and reported as mean values ± standard deviation, unless stated otherwise.
Acknowledgments
N.K. would like thank colleagues at BfR for their help and scientific input, in particular Chris Tina Höfer, Sebastian Dunst, Mariana Lara Neves, and Gilbert Schönfelder for critical comments on the manuscript. Furthermore, N.K. thanks Tra My Tran, as the first user of the software, for providing feedback and reporting bugs. Additionally, N.K. would like to thank Georges Hattab for his advice on the usability test and all the participants.
Author contributions
N.K. planned and designed the study, programmed the software, performed all experiments and analyses, and wrote the manuscript.
Declaration of interests
The author declares no competing interests.
Published: June 26, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2023.100517.
Supplemental information
Data and code availability
The complete source code of MIA can be found on https://github.com/MIAnalyzer/MIA with a user manual, installation options, and additional information. Additionally, the original source code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Krizhevsky A., Sutskever I., Hinton G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012 [Google Scholar]
- 2.Araújo T., Aresta G., Castro E., Rouco J., Aguiar P., Eloy C., Polónia A., Campilho A. Classification of breast cancer histology images using convolutional neural networks. PLoS One. 2017;12 doi: 10.1371/journal.pone.0177544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janowczyk A., Madabhushi A. Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J. Pathol. Inf. 2016;7:29. doi: 10.4103/2153-3539.186902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H., Rivenson Y., Jin Y., Wei Z., Gao R., Günaydın H., Bentolila L.A., Kural C., Ozcan A. Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat. Methods. 2019;16:103–110. doi: 10.1038/s41592-018-0239-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krull A., Buchholz T.-O., Jug F. Noise2void-learning denoising from single noisy images. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2019 [Google Scholar]
- 6.Weigert M., Schmidt U., Boothe T., Müller A., Dibrov A., Jain A., Wilhelm B., Schmidt D., Broaddus C., Culley S., et al. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods. 2018;15:1090–1097. doi: 10.1038/s41592-018-0216-7. [DOI] [PubMed] [Google Scholar]
- 7.Ounkomol C., Seshamani S., Maleckar M.M., Collman F., Johnson G.R. Label-free prediction of three-dimensional fluorescence images from transmitted-light microscopy. Nat. Methods. 2018;15:917–920. doi: 10.1038/s41592-018-0111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivenson Y., Wang H., Wei Z., de Haan K., Zhang Y., Wu Y., Günaydın H., Zuckerman J.E., Chong T., Sisk A.E., et al. Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat. Biomed. Eng. 2019;3:466–477. doi: 10.1038/s41551-019-0362-y. [DOI] [PubMed] [Google Scholar]
- 9.Bannon D., Moen E., Schwartz M., Borba E., Kudo T., Greenwald N., Vijayakumar V., Chang B., Pao E., Osterman E., et al. DeepCell Kiosk: scaling deep learning--enabled cellular image analysis with Kubernetes. Nat. Methods. 2021;18:43–45. doi: 10.1038/s41592-020-01023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ouyang W., Mueller F., Hjelmare M., Lundberg E., Zimmer C. ImJoy: an open-source computational platform for the deep learning era. Nat. Methods. 2019;16:1199–1200. doi: 10.1038/s41592-019-0627-0. [DOI] [PubMed] [Google Scholar]
- 11.von Chamier L., Laine R.F., Jukkala J., Spahn C., Krentzel D., Nehme E., Lerche M., Hernández-Pérez S., Mattila P.K., Karinou E., et al. Democratising deep learning for microscopy with ZeroCostDL4Mic. Nat. Commun. 2021;12:2276. doi: 10.1038/s41467-021-22518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falk T., Mai D., Bensch R., Çiçek Ö., Abdulkadir A., Marrakchi Y., Böhm A., Deubner J., Jäckel Z., Seiwald K., et al. U-Net: deep learning for cell counting, detection, and morphometry. Nat. Methods. 2019;16:67–70. doi: 10.1038/s41592-018-0261-2. [DOI] [PubMed] [Google Scholar]
- 13.Berg S., Kutra D., Kroeger T., Straehle C.N., Kausler B.X., Haubold C., Schiegg M., Ales J., Beier T., Rudy M., et al. Ilastik: interactive machine learning for (bio) image analysis. Nat. Methods. 2019;16:1226–1232. doi: 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- 14.McQuin C., Goodman A., Chernyshev V., Kamentsky L., Cimini B.A., Karhohs K.W., Doan M., Ding L., Rafelski S.M., Thirstrup D., et al. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen C., Miura T., Voleti V., Yamaguchi K., Tsutsumi M., Yamamoto K., Otomo K., Fujie Y., Teramoto T., Ishihara T., et al. 3DeeCellTracker, a deep learning-based pipeline for segmenting and tracking cells in 3D time lapse images. Elife. 2021;10 doi: 10.7554/eLife.59187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gómez-de-Mariscal E., García-López-de-Haro C., Ouyang W., Donati L., Lundberg E., Unser M., Muñoz-Barrutia A., Sage D. DeepImageJ: a user-friendly environment to run deep learning models in ImageJ. Nat. Methods. 2021;18:1192–1195. doi: 10.1038/s41592-021-01262-9. [DOI] [PubMed] [Google Scholar]
- 17.Stringer C., Wang T., Michaelos M., Pachitariu M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods. 2021;18:100–106. doi: 10.1038/s41592-020-01018-x. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt U., Weigert M., Broaddus C., Myers G. Springer; 2018. Cell Detection with Star-Convex Polygons; pp. 265–273. [Google Scholar]
- 19.Haberl M.G., Churas C., Tindall L., Boassa D., Phan S., Bushong E.A., Madany M., Akay R., Deerinck T.J., Peltier S.T., Ellisman M.H. CDeep3M—plug-and-Play cloud-based deep learning for image segmentation. Nat. Methods. 2018;15:677–680. doi: 10.1038/s41592-018-0106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollandi R., Szkalisity A., Toth T., Tasnadi E., Molnar C., Mathe B., Grexa I., Molnar J., Balind A., Gorbe M., et al. nucleAIzer: a parameter-free deep learning framework for nucleus segmentation using image style transfer. Cell Syst. 2020;10:453–458.e6. doi: 10.1016/j.cels.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann B., Walter T., Hériché J.K., Bulkescher J., Erfle H., Conrad C., Rogers P., Poser I., Held M., Liebel U., et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandler M., Howard A., Zhu M., Zhmoginov A., Chen L.-C. Mobilenetv2: inverted residuals and linear bottlenecks. Proceedings of the IEEE conference on computer vision and pattern recognition. 2018 [Google Scholar]
- 24.Zoph B., Vasudevan V., Shlens J., Le Q.V. Learning transferable architectures for scalable image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition. 2018 [Google Scholar]
- 25.Ehteshami Bejnordi B., Veta M., Johannes van Diest P., Van Ginneken B., Karssemeijer N., Litjens G., van der Laak J.A.W.M., the CAMELYON16 Consortium. Hermsen M., Manson Q.F., et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veeling B.S., Linmans J., Winkens J., Cohen T., Welling M. Springer; 2018. Rotation Equivariant CNNs for Digital Pathology; pp. 210–218. [Google Scholar]
- 27.Edlund C., Jackson T.R., Khalid N., Bevan N., Dale T., Dengel A., Ahmed S., Trygg J., Sjögren R. LIVECell—a large-scale dataset for label-free live cell segmentation. Nat. Methods. 2021;18:1038–1045. doi: 10.1038/s41592-021-01249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rother C., Kolmogorov V., Blake A. GrabCut" interactive foreground extraction using iterated graph cuts. ACM Trans. Graph. 2004;23:309–314. [Google Scholar]
- 29.Xie S., Tu Z. Holistically-nested edge detection. Proceedings of the IEEE international conference on computer vision. 2015 [Google Scholar]
- 30.Maninis K.-K., Caelles S., Pont-Tuset J., Van Gool L. Deep extreme cut: from extreme points to object segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2018 [Google Scholar]
- 31.Ronneberger O., Fischer P., Brox T. U-net: convolutional networks for biomedical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention. 2015 [Google Scholar]
- 32.Chen L.-C., Zhu Y., Papandreou G., Schroff F., Adam H. Encoder-decoder with atrous separable convolution for semantic image segmentation. Proceedings of the European Conference on Computer Vision (ECCV) 2018 [Google Scholar]
- 33.Tan M., Le Q. International Conference on Machine Learning; 2019. Efficientnet: Rethinking Model Scaling for Convolutional Neural Networks. [Google Scholar]
- 34.Kuhn H.W. The Hungarian method for the assignment problem. Nav. Res. Logist. 1955;2:83–97. [Google Scholar]
- 35.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M., et al. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015;115:211–252. [Google Scholar]
- 36.Kingma D.P., Ba J. Adam: a method for stochastic optimization. arXiv. 2014 doi: 10.48550/arXiv.1412.6980. Preprint at. [DOI] [Google Scholar]
- 37.Lin T.-Y., Goyal P., Girshick R., He K., Dollár P. Focal loss for dense object detection. Proceedings of the IEEE international conference on computer vision. 2017 doi: 10.1109/TPAMI.2018.2858826. [DOI] [PubMed] [Google Scholar]
- 38.Ruggieri A., Dazert E., Metz P., Hofmann S., Bergeest J.-P., Mazur J., Bankhead P., Hiet M.-S., Kallis S., Alvisi G., et al. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe. 2012;12:71–85. doi: 10.1016/j.chom.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulman V., Maška M., Magnusson K.E.G., Ronneberger O., Haubold C., Harder N., Matula P., Matula P., Svoboda D., Radojevic M., et al. An objective comparison of cell-tracking algorithms. Nat. Methods. 2017;14:1141–1152. doi: 10.1038/nmeth.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Svoboda D., Ulman V. MitoGen: a framework for generating 3D synthetic time-lapse sequences of cell populations in fluorescence microscopy. IEEE Trans. Med. Imaging. 2017;36:310–321. doi: 10.1109/TMI.2016.2606545. [DOI] [PubMed] [Google Scholar]
- 41.Chollet F. Xception: deep learning with depthwise separable convolutions. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017 [Google Scholar]
- 42.Szegedy C., Ioffe S., Vanhoucke V., Alemi A.A. Thirty-first AAAI conference on artificial intelligence; 2017. Inception-v4, Inception-Resnet and the Impact of Residual Connections on Learning. [Google Scholar]
- 43.Martín A., Ashish A., Paul B., Eugene B., Zhifeng C., Craig C., Greg S.C., Andy D., Jeffrey D., Matthieu D., et al. 2015. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems. [Google Scholar]
- 44.Wilkinson M.D., Dumontier M., Aalbersberg I.J.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E., et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016;3:160018–160019. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris C.R., Millman K.J., van der Walt S.J., Gommers R., Virtanen P., Cournapeau D., Wieser E., Taylor J., Berg S., Smith N.J., et al. Array programming with NumPy. Nature. 2020;585:357–362. doi: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bradski G. Dr. Dobb's Journal of Software Tools; 2000. The OpenCV Library. [Google Scholar]
- 47.van der Walt S., Schönberger J.L., Nunez-Iglesias J., Boulogne F., Warner J.D., Yager N., Gouillart E., Yu T., scikit-image contributors scikit-image: image processing in Python. PeerJ. 2014;2:e453. doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jung A.B. GitHub repository; 2018. Imgaug. [Google Scholar]
- 49.Chollet F., et al. GitHub repository; 2015. Keras. [Google Scholar]
- 50.keras t. GitHub repository; 2019. Keras-Applications. [Google Scholar]
- 51.Yakubovskiy P. GitHub repository; 2019. Segmentation Models. [Google Scholar]
- 52.Yakubovskiy P. GitHub repository; 2019. Classification Models. [Google Scholar]
- 53.Zakirov E. GitHub repository; 2019. keras-deeplab-v3-plus. [Google Scholar]
- 54.Nvidia Vingelmann P., Fitzek F.H.P. 2020. CUDA. [Google Scholar]
- 55.Beucher S. The watershed transformation applied to image segmentation. Scanning Microsc. 1992;1992:28. [Google Scholar]
- 56.Brooke J. SUS: a quick and dirty usability scale. Usability Eval. Ind. 1995;189 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete source code of MIA can be found on https://github.com/MIAnalyzer/MIA with a user manual, installation options, and additional information. Additionally, the original source code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.