Summary
O-linked N-acetylglucosaminylation (O-GlcNAcylation) is a ubiquitous and dynamic non-canonical glycosylation of intracellular proteins. Several branches of metabolism converge at the hexosamine biosynthetic pathway (HBP) to produce the substrate for protein O-GlcNAcylation, the uridine diphosphate N-acetylglucosamine (UDP-GlcNAc). Availability of UDP-GlcNAc is considered a key regulator of O-GlcNAcylation. Yet UDP-GlcNAc concentrations are rarely reported in studies exploring the HBP and O-GlcNAcylation, most likely because the methods to measure it are restricted to specialized chromatographic procedures. Here, we introduce an enzymatic method to quantify cellular and tissue UDP-GlcNAc. The method is based on O-GlcNAcylation of a substrate peptide by O-linked N-acetylglucosamine transferase (OGT) and subsequent immunodetection of the modification. The assay can be performed in dot-blot or microplate format. We apply it to quantify UDP-GlcNAc concentrations in several mouse tissues and cell lines. Furthermore, we show how changes in UDP-GlcNAc levels correlate with O-GlcNAcylation and the expression of OGT and O-GlcNAcase (OGA).
Keywords: hexosamine biosynthetic pathway, O-linked N-acetylglucosamine transferase, OGT, O-GlcNAcase, OGA, nucleotide sugars, microplate assay for UDP-GlcNAc, protein O-GlcNAcylation homeostasis, GlcNAc salvage pathway
Graphical abstract

Highlights
-
•
We developed an OGT- and O-GlcNAc antibody-based assay for UDP-GlcNAc
-
•
The assay can be performed in dot-blot or microplate format
-
•
Cellular protein O-GlcNAcylation is resistant to acute changes in UDP-GlcNAc levels
-
•
OGT-to-OGA ratio serves as a sensitive indicator of UDP-GlcNAc levels
Motivation
Current methods to quantify uridine diphosphate N-acetylglucosamine (UDP-GlcNAc), the end product of the hexosamine biosynthetic pathway and the substrate for protein O-GlcNAcylation, involve specialized chromatographic techniques. These methods are not readily available to all research laboratories. Moreover, the identical molecular mass and the near identical reactivity and elution profile of N-acetylhexosamine epimers make their separate quantification challenging. To overcome these limitations, we developed a sensitive and practical high-throughput enzymatic assay for UDP-GlcNAc.
Sunden et al. present an assay for UDP-GlcNAc, the end product of the hexosamine biosynthetic pathway and the substrate for protein O-GlcNAcylation. The assay can be performed with commercially available reagents in any laboratory harboring a typical plate reader. The sample extraction presented enables parallel quantification of UDP-GlcNAc, O-GlcNAc cycling enzymes, and protein O-GlcNAcylation.
Introduction
The hexosamine biosynthetic pathway (HBP) consumes uridine triphosphate (UTP), glucose, glutamine, and acetyl-CoA to produce two N-acetylated amino sugars coupled to uridine diphosphate (UDP): UDP-N-acetylgalactosamine (UDP-GalNAc) and UDP-N-acetylglucosamine (UDP-GlcNAc).1 In addition to being a biosynthetic precursor for glycan chains of the secretory pathway and extracellular proteins, UDP-GlcNAc is the substate for the monomeric O-linked N-acetylglucosaminylation (O-GlcNAcylation) of serine and threonine residues of intracellular proteins. The sole enzyme performing this specific glycosylation in mammalian cells is the O-linked N-acetylglucosamine transferase (OGT). Likewise, only one enzyme, the O-GlcNAcase (OGA), removes this modification. Although it is a seemingly simple two-enzyme regulation, an increasing number of studies showing altered protein O-GlcNAcylation in various pathological conditions and experimental models implicate O-GlcNAc as a highly dynamic protein post-translational modification.1
Many reviews have postulated UDP-GlcNAc as the central regulatory metabolite governing protein O-GlcNAcylation.2,3,4,5 Indeed, the HBP integrates nucleotide, carbohydrate, amino acid, and fatty acid metabolism, making UDP-GlcNAc a sensible regulatory node for energy metabolism. Nevertheless, cellular or tissue concentrations of UDP-GlcNAc have rarely been reported, even in many seminal studies on O-GlcNAcylation. On the basis of limited literature, tissue UDP-GlcNAc concentrations range from 10 to 35 μM in the skeletal muscle to ∼150 μM in the liver,6,7,8,9,10 while OGT requires only 0.5–5 μM UDP-GlcNAc for its half-maximal activity,11,12,13 contradicting the UDP-GlcNAc availability as a sensitive regulator of OGT activity at least in some tissues. However, UDP-GlcNAc does not equally distribute within a cell but is actively concentrated into the endoplasmic reticulum and Golgi apparatus.14 Unfortunately, little is known about the effective UDP-GlcNAc concentration in different cellular compartments.
Traditional methods to measure nucleotide sugars involve chromatographic procedures such as liquid chromatography or capillary electrophoresis.10,15,16 These methods require expensive special equipment and expertise, and thus are not easily applicable in a typical life science research laboratory. Moreover, the near identical chromatographic elution profile, reactivity, physical properties, and the identical molecular mass of the nucleotide sugar epimers, make their separate quantification (e.g., UDP-GlcNAc from UDP-GalNAc) challenging using these methods.17 For these reasons, UDP-GlcNAc and UDP-GalNAc concentrations are typically reported as a sum of these two metabolites (UDP-N-acetylhexosamines [UDP-HexNAcs]). The biological functions of the two UDP-HexNAcs are, however, distinct.
To date, only one enzymatic assay for UDP-GlcNAc has been published.18 This assay is based on NAD+-dependent UDP-GlcNAc dehydrogenase from Methanococcus maripaludis. As the assay relies on direct monitoring of NADH generation, it is intrinsically limited in sensitivity. The authors showed, however, sufficient sensitivity to measure UDP-GlcNAc from yeast and HeLa cells. Nevertheless, to our knowledge, no further use of this method has been reported during the 14 years since its publication.
Here, we harnessed the high affinity of OGT for UDP-GlcNAc to develop an enzymatic assay for UDP-GlcNAc. We used the assay to measure UDP-GlcNAc concentrations in different mouse tissues and cultured cells. Finally, we examined the relationships among UDP-GlcNAc levels, protein O-GlcNAcylation, and the expression of OGT and OGA.
Results
Development of enzymatic assay for UDP-GlcNAc
After unsuccessful attempts to quantify UDP-GlcNAc with the previously reported enzymatic assay18 (Figure S1), we looked into methods to measure glycosyltransferase activity of OGT19,20 and whether these methods could be converted for the measurement of UDP-GlcNAc. Given the reported11 high affinity of OGT for UDP-GlcNAc and multiple options to detect O-GlcNAcylated proteins, we reasoned that OGT-mediated O-GlcNAcylation could, in theory, be used to develop an assay for UDP-GlcNAc. For proof-of-principle experiments, we set up a simple assay scheme involving human recombinant OGT fragment, a GlcNAc-acceptor peptide crosslinked to BSA, limiting concentrations of UDP-GlcNAc, and dot blotting onto PVDF membrane to capture the peptide-BSA complex for immunodetection of O-GlcNAcylated residues (Figure 1A). The acceptor peptide derives from human casein kinase 2 and is one of the most efficient substrates for O-GlcNAcylation reported.12 We reasoned that for the assay to work, we would need to remove interference by UDP, especially from biological samples. UDP is a reaction product in O-GlcNAcylation and a potent inhibitor (half maximal inhibitory concentration [IC50] < 1 μM) of OGT.11,21 To do this, we included alkaline phosphatase in the assay reactions. The pyrophosphate group in free nucleotides is exposed to hydrolysis by phosphatases, whereas in UDP-GlcNAc it is not as it is protected by the glucosamine and ribose moieties. For immunodetection of O-GlcNAc, we chose the mouse monoclonal antibody RL2.22 Although several other GlcNAc-specific antibodies and lectins exist, many of them cross-react with glycosylated proteins in blocking reagents such as skimmed milk and BSA or tend to give high background for other reasons. This is our experience and is also reported in the literature.19,23
Figure 1.
Enzymatic assay for UDP-GlcNAc with dot-blot detection
(A) A schematic presentation of the assay principle.
(B) A representative detection of O-GlcNAcylated peptides by dot blotting, RL2 monoclonal antibody, and chemiluminescence.
(C) Standard curve showing UDP-GlcNAc concentration-dependent increase in signal.
(D) Standard curve covering 0–1.25 μM UDP-GlcNAc after extended camera exposure time.
(E) Measured UDP-GlcNAc amount from a liver extract with and without addition of 20 pmol exogenous analyte.
(F) Effect of UDP degradation (by alkaline phosphatase) on measured signal from liver extract and purified UPD-GlcNAc standard sample. Liver extracts worth 144 μg pre-extraction tissue weight were used in 10 μL assay reactions. The error bars represent SEM of three technical replicates.
The initial assay concept was successful with purified UDP-GlcNAc standards and, importantly, with liver tissue extract (Figures 1B–1E). In the absence of OGT, the liver extract did not give any measurable signal, verifying complete removal of endogenous O-GlcNAcylated proteins (Figure 1B). Spiking the liver extract with a known amount of UDP-GlcNAc led to a corresponding increase in signal (Figure 1E), excluding major sample-derived interference. In line with our hypothesis, the inclusion of alkaline phosphatase proved to be indispensable for the assay (Figure 1F).
Enzymatic UDP-GlcNAc assay in microplate format
Although we found the dot-blot format simple, quantitative, and relatively sensitive, our charge-coupled device (CCD) camera-based imaging system limited the dynamic range. Moreover, this assay format requires a dot-blotting apparatus for quantitative results, a relatively rare piece of equipment nowadays. Because of these limitations, we modified the assay into 384-well microplate format (Figure 2A). We coated the wells of high protein-binding microplates with the GlcNAc-acceptor peptide-BSA complex. After blocking unoccupied protein-binding sites, we performed the O-GlcNAcylation reactions in the wells and used a typical direct ELISA-like detection using RL2 antibody and peroxidase-conjugated secondary antibody. We chose to develop the signal using Amplex UltraRed as the peroxidase substrate, giving a highly sensitive chemifluorescent readout. Figure 2B shows a standard curve generated from the endpoint fluorescence values.
Figure 2.
Enzymatic UDP-GlcNAc assay in microplate format
(A) A schematic presentation of the microplate assay for UDP-GlcNAc.
(B) Standard curve generated from endpoint fluorescence values.
(C) Comparison of chloride and acetate as counter anions in Bis-Tris-based assay buffer.
(D) Comparison of two phosphatases with different pH optimum on signal from liver extract. The assay buffer pH was 7.0.
(E) Effect of O-GlcNAcylation reaction temperature on the assay performance.
(F) Effect of reaction time on the sensitivity of the assay.
(G) Effect of reaction temperature with overnight incubations on the assay.
(H) Verification of the specificity with a biological sample by omission of OGT from the assay reactions.
(I) Effect of BSA in assay reactions on sample-related inhibition as assessed by spiking the samples with a known amount of UDP-GlcNAc. OGT concentration in these measurements was 10 μg/mL.
(J) Dispensability of the separate blocking step.
(K) Effect of desiccation of the coated plate.
(L) Comparison of human casein kinase II and the peptide-BSA complex as GlcNAc acceptors in the assay reactions.
(M) Suitability of three different antibodies for the detection step of the assay. CST, Cell Signaling Technology.
(N) A representative standard curve of the optimized assay.
(O) Correlation between UDP-GlcNAc (microplate assay) and UDP-HexNAc levels (UPLC-MS) in mouse liver samples. Bar graphs present mean and SEM of technical triplicates. All incubations steps were performed at room temperature (21°C–23°C) unless otherwise indicated in the figures.
Optimization of the assay
Using the dot-blot and microplate formats of the assay, we set out to optimize the assay parameters for highest performance. The reported pH optimum for OGT activity lies between 6 and 7.5.11 Different buffers (Tris-Cl, Bis-Tris-Cl, Bis-Tris-acetate, and HEPES-K [pH 7–7.5]) did not have a large impact on OGT activity under limiting substrate concentration (Figures 2C and S2A–S2C). OGT is highly sensitive to inhibition by potassium and sodium chloride according to some reports.11,21 We tested a HEPES-based buffer with a near physiological concentration of potassium (∼90 mM) but without unphysiological chloride content. This buffer was compatible with the assay but inferior to plain Bis-Tris buffer that lacked added salt (Figure S2B). When using Bis-Tris buffer, acetate proved an inferior counter anion in comparison with chloride (Figure 2C). For further experiments, we settled for 50 mM Bis-Tris (Cl−) buffer with pH 7.0. OGT does not require divalent cations for its activity,24 and, indeed, omission of Mg2+ had only a minor effect when assessing purified UDP-GlcNAc samples (Figure S1A). However, we decided to include 5 mM Mg2+ to maximize alkaline phosphatase activity, which might become limiting when assessing complex biological samples. The selected pH of 7.0 is not optimal for the alkaline phosphatase activity. However, a phosphatase with pH optimum close to 7 (Antarctic phosphatase; New England BioLabs) performed worse than alkaline phosphatase when assessing liver extracts (Figure 2D).
For the aforementioned experiments, we used a reaction time of 2 h at room temperature (21°C–23°C). OGT has been reported to be relatively stable in vitro at temperatures below 30°C but to undergo rapid inactivation at 37°C.21 In the dot-blot format, increase of reaction temperature to 37°C had only a minor buffer-dependent effect on the signal (Figure S1A). In the microplate format, however, the increase of reaction temperature to 37°C notably compromised the assay sensitivity (Figure 2E). This assay format difference suggests that GlcNAc-acceptor peptides, which were in excess and in solution in the dot-blot format, help stabilize OGT. We found that the GlcNAcylation reactions could be left to proceed overnight (∼16 h) at room temperature for convenience (Figure 2F). However, the extended reaction time increased both the specific signal and the background, therefore decreasing sensitivity (Figure 2F). Decrease of the temperature to 8°C for overnight reactions somewhat increased the sensitivity, whereas elevated temperature (37°C) had an opposite effect (Figure 2G).
Next, we assessed cell and liver extracts with the microplate format. Cell extracts gave measurable signal only when the assay reactions included OGT (Figure 2H), verifying the specificity. Surprisingly, the microplate format turned out to be more sensitive to sample-related inhibition than the dot-blot format (Figure 2I). In the dot-blot format, the reactions contained BSA (as a carrier for the GlcNAc-acceptor peptide). Therefore, we suspected that BSA might mitigate the sample-related inhibition. Indeed, this was the case (Figure 2I).
Because of the relatively high protein concentration (BSA, OGT, and alkaline phosphatase) in O-GlcNAcylation reactions and the ability of Tween 20 to serve as efficient blocking agent in many ELISA assay,25 we tested the dispensability of the separate blocking step (0.7% Na-caseinate and 1% BSA). This step turned out to be unnecessary (Figure 2J), and we omitted it from the final protocol.
For initial experiments, we used sodium carbonate bicarbonate buffer pH 9.6 to coat the microplates, but PBS worked equally well as the coating buffer (Figure S2D). The coated plates did not tolerate desiccation for storage without loss of the assay sensitivity (Figure 2K). As an alternative GlcNAc acceptor, we tested coating with commercial human casein kinase 2 (composed of both α- and β-subunits). This GlcNAc acceptor gave essentially identical assay behavior to the peptide-BSA complex when UDP-GlcNAc concentrations were below 125 nM (Figure 2L). At higher concentrations, the signal plateaued earlier than with the peptide-BSA complex, suggesting that the GlcNAcylation sites became limiting. Next, we compared the RL2 antibody against another widely used mouse monoclonal antibody against O-GlcNAc the CTD110.6 and a more recent rabbit monoclonal antibody mixture from Cell Signaling Technology (catalog #82332). The O-GlcNAc antibody mixture from Cell Signaling Technology was unsuitable for the assay (Figure 2M). CTD110.6 gave higher background than RL2 (Figure S2E), and therefore lower sensitivity (Figure 2M), but the practical difference between these two antibodies was quite negligible.
Finally, we tested the lowest limit of quantification (LLOQ) of the optimized assay (Figure 2N). Our microplate format reached the LLOQ of 110 fmol (5.5 nM UDP-GlcNAc in reaction). The estimated lowest limit of detection (LLOD) was 40 fmol.
UDP-GlcNAc concentrations in mouse tissues
To test the applicability of our method, we measured UDP-GlcNAc from several mouse tissues. Different nucleotide sugars extraction procedures have been reported in the literature ranging from acid (e.g., perchloric acid and trichloroacetic acid) to solvent extractions.10,17,18 We chose MeOH-H2O-CHCl3 extraction, which has been shown to give close to 100% recovery for nucleotide sugars,26 and which we found compatible with the enzymatic method and ultra-performance liquid chromatography-mass spectrometry (UPLC-MS). For this extraction method to be compatible with the enzymatic method, we removed the residual MeOH from the aqueous phase by repeated CHCl3 or diethyl ether washes, or alternatively by drying the extracts. The recovery of exogenous UDP-GlcNAc spiked to kidney biopsies immediately after the tissue disruption was more than 90% (Figure S3). As an assay validation, we measured UDP-GlcNAc from 23 mouse liver samples from which we had existing UDP-HexNAc data (determined using UPLC-MS). Assuming unrestricted cellular epimerization between UDP-GlcNAc and UDP-GalNAc, the levels of these two metabolites should show high correlation. This was indeed the case, and the UDP-GlcNAc concentrations strongly correlated with the total UDP-HexNAc levels (Figure 2O).
Table 1 shows UDP-GlcNAc concentrations in the mouse liver, kidney, heart, skeletal muscle, and brain. Of these tissues, the liver had the highest UDP-GlcNAc concentration (240 pmol/mg). This concentration was of similar magnitude to previously reported values (125–150 pmol/mg).6 The skeletal muscle contained the smallest amount of UDP-GlcNAc (14 pmol/mg). Our estimate was very close to a previously reported concentration in mouse skeletal muscle (11 pmol/mg)6 and of similar magnitude to that reported in different skeletal muscle tissues in rat (25–36 nmol/g).7,9,10 In another muscle tissue, in the heart, UDP-GlcNAc content was somewhat higher, 42 pmol/mg. Previous studies have shown that UDP-HexNAc content in the mouse and rat heart is roughly in the range of 40–60 pmol/mg,27,28 and most of this is UDP-GlcNAc.28 In the rat brain, UDP-HexNAc content has been reported to be approximately 130 pmol/mg. Our estimate of UDP-GlcNAc concentration in mouse brain tissue was 62 pmol/mg.
Table 1.
UDP-GlcNAc concentration in mouse tissues (pmol/mg tissue)
| Tissue | Mean | SD | Sample input, μg initial tissue mass | Recovery of 0.5 pmol exogenous UDP-GlcNAc, % |
|---|---|---|---|---|
| Liver | 241 | 57 | 3.4 | 109 |
| Kidney | 145 | 37 | 4.6 | 104 |
| Heart | 41.6 | 9.3 | 9.5 | 117 |
| Skeletal muscle (quadriceps) | 13.7 | 0.8 | 63.5 | 102 |
| Brain (cerebrum) | 62.6 | 7.4 | 9.9 | 135 |
Brain, n = 3 mice; all other tissues, n = 4 mice. Exogenous UDP-GlcNAc was spiked into replicate final extracts.
UDP-GlcNAc levels in cultured mammalian cells
Table 2 lists cellular UDP-GlcNAc content in 7 different cell lines: 293T, NIH/3T3, HCT116, AML12, Hepa1-6, HeLa cells, and primary mouse fibroblasts. The UDP-GlcNAc concentrations ranged from 60 to 520 pmol/million cells, with the highest per cell content in HeLa cells, followed by the liver-originating cells.
Table 2.
UDP-GlcNAc concentration in cultured mammalian cells (pmol/106 cells)
| Cell line | Cell type/origin | Mean | SD |
|---|---|---|---|
| 293T | immortalized epithelial-like cells from human embryonic kidney | 134 | 42 |
| NIH/3T3 | spontaneously immortalized mouse embryonic fibroblasts | 64 | 2.6 |
| Mouse skin fibroblasts | primary cells | 102 | 16 |
| HCT116 | human colorectal carcinoma | 120 | 25 |
| AML12 | mouse hepatocytes transgenic for human TGF-α | 220 | 56 |
| Hepa 1-6 | mouse hepatoma | 160 | 35 |
| HeLa | human cervical carcinoma | 520 | 160 |
n = 3 cell culture flasks per cell line.
The HBP and protein O-GlcNAcylation have been targeted by various indirect and direct approaches in published studies, but data on the effect of these manipulations on UDP-GlcNAc levels remain surprisingly scarce. Therefore, we assessed UDP-GlcNAc levels in AML12 hepatocyte cell line subjected to different culture conditions and metabolic stressors (Figure 3A). Cells grown in 5 or 25 mM glucose had similar UDP-GlcNAc levels, which is coherent with observations that excess glucose does not significantly increase UDP-GlcNAc levels in HepG2 hepatocellular carcinoma cells,29,30 3T3-L1 adipocytes,31 or perfused mouse heart.28 In contrast, complete withdrawal of glucose for 16 h decreased the UDP-GlcNAc levels by 65%. This depletion was partially prevented by 1 mM N-acetylglucosamine (GlcNAc) but not by galactose supplementation. In the presence of glucose, GlcNAc elevated the UDP-GlcNAc levels above that of standard culture conditions. Serum-free medium increases hepatocyte-specific gene expression without blocking proliferation in AML12 cells.32 Upon serum deprivation, AML12 cells increased their UDP-GlcNAc content (Figures 3A and 3B).
Figure 3.
Relationships among UDP-GlcNAc content, protein O-GlcNAcylation, and the expression of OGT and OGA in AML12 cells
(A) UDP-GlcNAc concentration and OGT-to-OGA ratio (western blot) in cells subjected to the indicated culturing conditions for approximately 16 h. Figure S4 shows representative Western blot detections of OGT, OGA, and O-GlcNAcylated proteins.
(B) Effect of serum deprivation on UDP-GlcNAc levels.
(C) Effect of GFAT inhibition (azaserine) on UDP-GlcNAc levels.
(D) Western blot quantification of protein O-GlcNAcylation, and the expression of OGT and OGA. The samples are same as in (A).
(E) Correlations between UDP-GlcNAc concentration and the amount of O-GlcNAcylated proteins, and the expression of OGT and OGA, and the OGT-to-OGA ratio. The data points represent an average of three replicate cell culture dishes from data in (A) and (D). The small red numerals refer to the experimental groups in (A) and (D), ordered from left to right.
Data from GlcNAc-supplemented cells (blue and red triangles) were not included in the regression analysis. ∗p < 0.05 (1-way ANOVA followed by Dunnett’s test, 25 mM glucose as control). In (B) and (C), an unpaired two-sided t test was used. Error bars represent ±SD.
Azaserine is a widely used, albeit rather unspecific, inhibitor of the HBP. It inhibits glutamine-dependent enzymes including glutamine fructose-6-phosphate amidotransferases (GFPTs; previously called GFATs) the rate-limiting enzymes of the HBP. In AML12 cells, azaserine decreased UDP-GlcNAc by 50% (Figure 3C), which is similar to that observed in three endothelial cell lines.33,34,35
Parallel quantification of UDP-GlcNAc, O-GlcNAcylation, and expression of OGT and OGA
Our metabolite extraction procedure allows parallel collection of the total protein fraction. We took advantage of this possibility to measure UDP-GlcNAc and protein O-GlcNAcylation from the same samples. Despite the clear difference in UDP-GlcNAc concentration, the acute (∼16 h) changes in protein O-GlcNAcylation in the manipulated AML12 cells were minimal (Figures 3A, 3D, 3E, and S4). Cells have been reported to compensate protein O-GlcNAcylation disturbances by rapidly regulating the expression of OGT and OGA.29,36 Here, we analyzed the expression patterns of these two enzymes in relation to the UDP-GlcNAc concentration in AML12 cells (Figures 3A, 3D, 3E, and S4). The OGT expression negatively correlated with the cellular UDP-GlcNAc, while the reverse was true for OGA (Figure 3E). The changes were relatively small but given the opposite direction of change, the OGT-to-OGA ratio correlated strongly with the cellular UDP-GlcNAc content. An exception to this correlation came from cells cultured in the presence of 1 mM GlcNAc.
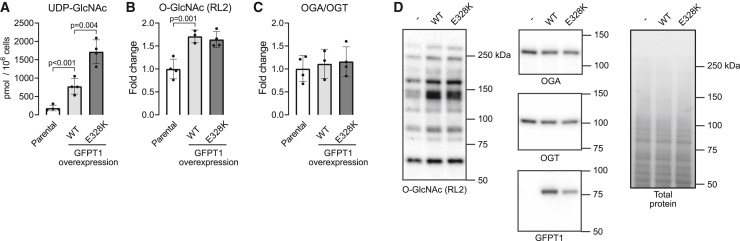
In order to examine the consequences of supraphysiological UDP-GlcNAc levels without GlcNAc supplementation, we generated AML12 cell lines constitutively overexpressing either wild-type (WT) or E328K gain-of-function mutant37 GFPT1, an enzyme catalyzing the rate-limiting step of the HBP. Overexpression of the WT enzyme increased UDP-GlcNAc levels approximately 5-fold and the E328K mutant 10-fold (Figure 4A). These supraphysiological UDP-GlcNAc levels led to a relatively meager 1.7-fold increase in O-GlcNAcylated proteins (Figures 4B and 4D). OGA and OGT expression did not respond to the supraphysiological UDP-GlcNAc availability (Figures 4C and 4D), suggesting together with the data in Figure 3 that the altered expression of OGA and OGT balance O-GlcNAcylation due to diminished but not elevated UDP-GlcNAc levels in this cell line.
Figure 4.
Effect of hexosamine biosynthetic pathway hyperactivity on cellular UDP-GlcNAc and protein O-GlcNAcylation in AML12 cells
(A–C) UDP-GlcNAc content (A), protein O-GlcNAcylation (B), and OGA-to-OGT expression ratio (C) in parental cells and cells with stable overexpression of wild-type (WT) or E328K mutant GFPT1.
(D) Representative western blots and total protein staining as a loading control.
One-way ANOVA followed by the selected pairwise comparisons (t test). Data points represent replicate cell culture flasks. Error bars represent mean and ±SD.
Finally, we measured the same parameters as above from a pancreatic adenocarcinoma cell line (TU8988T) with biallelic GFPT1 knockout. These cells rely on GlcNAc salvage pathway for the generation of UPD-GlcNAc and survive only in the presence of exogenous source of GlcNAc.38 The parental and the knockout cells had similar very high UDP-GlcNAc content when grown in the presence of 10 mM GlcNAc (Figure 5A; note the logarithmic scale). After 24 h of GlcNAc withdrawal, the knockout cells had lost more than 90% of their UDP-GlcNAc, but they still continued to proliferate (Figures 5A and 5E). Decreases in O-GlcNAcylated proteins and OGA-to-OGT ratio paralleled the UDP-GlcNAc decline (Figures 5A–5D). After 48 h, the GlcNAc-starved knockout cells had only ∼1% of the baseline UDP-GlcNAc concentration and no detectable O-GlcNAcylated proteins, and notably decreased OGA-to-OGT ratio. On the third day of GlcNAc withdrawal, the OGA-to-OGT ratio had further decreased. Growth curves indicated gradual loss of viability after 3–4 days in the GlcNAc-free media (Figure 5E).
Figure 5.
Effect of disrupted hexosamine biosynthetic pathway on cellular UDP-GlcNAc and protein O-GlcNAcylation in a pancreatic adenocarcinoma cell line (TU8988T)
(A–C) UDP-GlcNAc content (A), protein O-GlcNAcylation (B), and OGA-to-OGT expression ratio (C) in parental and GFPT1 knockout cells with and without 10 mM GlcNAc in media. Note the logarithmic y axis in (A). The indicated time points refer to duration since replacement of the culture media (start of the GlcNAc starvation).
(D) Representative western blots and total protein staining.
(E) Growth curves of the GFPT1 knockout cells.
∗Bonferroni-corrected p < 0.0001 (1-way ANOVA followed by the selected pairwise comparisons). The data points represent replicate cell culture flasks. Error bars represent ±SD.
Discussion
Knowledge on alterations in cellular UDP-GlcNAc concentration is crucial for understanding the mechanisms of O-GlcNAcylation disturbances in different experimental and disease settings. More than 2000 original articles dealing with protein O-GlcNAcylation or the HBP are listed on PubMed, yet we were able to find only a handful of reports showing data on UDP-GlcNAc concentrations. This disproportionate lack of UDP-GlcNAc data is at least partly because, thus far, a simple, practical, and sensitive method to measure this metabolite has not existed. We herein established a robust technique to measure UDP-GlcNAc in any laboratory harboring a typical plate reader and common biochemistry and molecular biology tools. All the reagents are commercially available or, alternatively, the required recombinant enzymes can be produced in-house in bacteria using standard procedures. Moreover, using the sample extraction described here, it is possible to measure UDP-GlcNAc, protein O-GlcNAcylation, and the expression of OGT and OGA from the same samples.
Unexpectedly, we found that global protein O-GlcNAcylation is highly resistant to acute changes in UDP-GlcNAc levels. Only after rather extreme manipulations of the HBP flux or knockout or overexpression of GFPT1, we achieved a clear loss of O-GlcNAcylation homeostasis. These findings challenge the widely postulated concept of the HBP being a sensitive sensor of nutrient availability and protein O-GlcNAcylation being primarily regulated by substrate availability.2,3,4,5 Our findings are, however, in line with the observations that many cellular stressors, predicted to compromise cellular energy status, actually increase protein O-GlcNAcylation and that excess cellular glucose intake can have minimal or no effect on the HBP flux.28,31,39 Two potential reasons for the stability of protein O-GlcNAcylation are the high total cellular concentration of UDP-GlcNAc and the very high affinity of OGT for it.11,12,13 Nevertheless, the expression of OGT and OGA did change upon any decline in UDP-GlcNAc levels, implying rigorous cellular mechanism to sense the UDP-GlcNAc and O-GlcNAcylation statuses. In fact, the OGT-to-OGA ratio proved to be a very sensitive marker of decreased UDP-GlcNAc availability. In contrast, forced excess of UDP-GlcNAc had less robust effect on the expression of OGT and OGA, likely meaning that normal cellular UDP-GlcNAc concentrations are close to saturating for maximal OGT activity. One practical implication of these findings is that the OGT-to-OGA ratio may serve as a much better marker of decreased UDP-GlcNAc levels than the widely used global protein O-GlcNAcylation.
Chemical inhibition or knockdown of OGT leads to compensatory downregulation of OGA and vice versa,36,40,41,42 indicating mutual regulation of these enzymes by protein O-GlcNAcylation. Whether UDP-GlcNAc levels affect the expression of OGT and OGA solely via protein O-GlcNAcylation or also independently of it, remains yet to be clarified. Experiments performed with purified proteins suggest that the affinity of OGT for UDP-GlcNAc varies depending on the GlcNAc-acceptor protein.12 Thus, it is possible that O-GlcNAcylation of some signaling proteins regulating the two O-GlcNAc cycling enzymes is more sensitive to UDP-GlcNAc availability than global protein O-GlcNAcylation, perhaps explaining the strong correlation between OGT-to-OGA ratio and UDP-GlcNAc even without notable differences in global protein O-GlcNAcylation. Evidence also exists for O-GlcNAcylation-independent regulation of OGT and OGA levels. This evidence comes from studies on the paradoxical increase of protein O-GlcNAcylation upon glucose deprivation despite decreased UDP-GlcNAc levels in several cancer cell lines and primary cardiomyocytes.29,30,43,44,45 Counterintuitively, this response can be blocked with very modest supplementation with glucosamine (0.02–1 mM), which prevents downregulation of OGA and, depending on a cell line or experimental design, upregulation of OGT.30,45 In our experiments with the non-transformed AML12 hepatocyte cell line, glucose deprivation did not increase the amount of O-GlcNAcylated proteins. However, supplementation of the glucose-starved AML12 cells with GlcNAc, an acetylated form of glucosamine, distorted the correlation between OGT-to-OGA ratio and UDP-GlcNAc levels, suggesting that GlcNAc or its phosphorylated intermediates of the HBP (GlcNAc 1-phosphate or GlcNAc 6-phosphate) are involved in the regulation of OGA. Intriguingly, glucosamine supplementation modulates several signaling pathways regulating glucose metabolism.46
The relationships among the HBP flux, UDP-GlcNAc levels, the O-GlcNAc cycling enzymes, and protein O-GlcNAcylation remain insufficiently understood. We expect that our method will facilitate tracking down how alterations in cellular UDP-GlcNAc levels affect global and protein-specific O-GlcNAcylation in cell lines and in vivo. We also expect that the possibility to quantify UDP-GlcNAc in small biological samples easily will help unravel the underlying causes for altered O-GlcNAcylation in different experimental and disease settings.
In conclusion, we developed a sensitive microplate assay for UDP-GlcNAc, the end product of the HBP and substrate for protein O-GlcNAcylation. Although a large repertoire of tools to probe O-GlcNAcylation have emerged, assessing UDP-GlcNAc has remained difficult for non-specialized laboratories.47 Our assay fills this methodological gap.
Limitations of the study
The assay described here is highly sensitive, with an LLOQ of 110 fmol, meaning, e.g., that 1 mg tissue biopsy or 50,000 typical cultured cells will yield more than enough extract for the quantification of UDP-GlcNAc. Although not empirically tested, the assay can be considered free of interference by UDP-GalNAc because of the epimer-specific enzymatic reaction11,48 and epimer-specific immunodetection.22 However, there are some limitations and room for further improvement of the assay. Although the dynamic range with purified UDP-GlcNAc standards is wide, sample-related interference remains a possibility that should be assessed for new sample types if aiming for absolute quantification of UDP-GlcNAc. Luckily, the high sensitivity of the assay likely allows dilution of any interference encountered. Performing the assay takes a full workday. However, although not systematically compared here, all incubation steps can be left to proceed overnight for convenience with relatively minimal effect on the assay performance. One requirement for standardization of the assay would be determination of the optimal specific activity of OGT. For those interested in further development of the assay, the provided Table S1 lists some qualitative observations and rationale for the selected assay parameter choices.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Clone RL2 | Biolegend | Cat#677902; RRID:AB_2565912 |
| Clone CTD110.6 | Biolegend | Cat#838004; RRID:AB_2629520 |
| GFPT1 | Abcam | Cat#ab125069; clone EPR4854; RRID:AB_10975709 |
| OGT | Abcam | Cat#ab96718; RRID:AB_10680015 |
| OGA | Merck | Cat#HPA036141: RRID:AB_10672079 |
| α-tubulin | Cell Signaling Technology | Cat#3873; clone DM1A; RRID:AB_1904178 |
| Anti-mouse IgG peroxidase conjugate | Cell Signaling Technology | Cat#7076; RRID:AB_330924 |
| Anti-mouse IgM peroxidase conjugate | Jackson Immunoresearch | Cat#115-035-020; RRID:AB_2338502 |
| Anti-rabbit IgG peroxidase conjugate | Cell Signaling Technology | Cat#7074; RRID:AB_2099233 |
| Bacterial and virus strains | ||
| E. coli BL21 | in-lab | N/A |
| E. coli DH5alpha | in-lab | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Human Recombinant OGT | R&D systems | Cat#8446-GT |
| Alkaline phosphatase | Thermo Scientific | Cat#EF0651 |
| Antarctic phosphatase | New England BioLabs | Cat#M0289 |
| Human casein kinase II | New England BioLabs | Cat#P6010 |
| Benzonase | Sigma-Aldrich | Cat#E1014 |
| cOmplete™, EDTA-free Protease Inhibitor Cocktail | Roche | Cat#04693132001 |
| HisPur Cobalt resin | Thermo Scientific | Cat#89964 |
| GlcNAc-acceptor peptide (KKKYPGGSTPVSSANMM) | Nordic BioSite | Custom synthesis |
| Nunc® MaxiSorp™ 384 | Sigma-Aldrich | Cat#P6116 |
| UDP-GlcNAc | Sigma-Aldrich | Cat#U4375 |
| Amplex UltraRed | Invitrogen | Cat#A36006 |
| Experimental models: Cell lines | ||
| AML12 mouse hepatocyte cell line | ATCC | Cat#CRL-2254 |
| AML12 cells overexpression wild-type GFPT1 | This paper | N/A |
| AML12 cells overexpressing E328K mutant GFPT1 | This paper | N/A |
| TU8988T pancreatic adenocarcinoma cell line | Kim et al. 202138 | N/A |
| GFPT1 knockout TU8988T cells | Kim et al. 202138 | N/A |
| HCT 116 Homo human colorectal carcinoma | ATCC | Cat#CCL-247 |
| NIH/3T3 mouse embryonic fibroblast | ATCC | Cat#CRL-1658 |
| Hepa1-6 mouse hepatoma | ATCC | Cat#CRL-1830 |
| 293T human kidney epithelium | ATCC | Cat#CRL-3216 |
| HeLa human adenocarcinoma | ATCC | Cat#CRM-CCL-2 |
| Experimental models: Organisms/strains | ||
| Congenic C57BL/6JCrl mice | Harlan | RRID:IMSR_JAX:000664 |
| Oligonucleotides | ||
| Human OGT PCR cloning MunI-OGT 5′-ATCAATTGACCA TGCTGTGTCCCACCCATGCAGA-3′ |
Metabion | N/A |
| Human OGT PCR cloning: OGT-STP-SalI 5′-ATATGTCGAC TTATTCAACAGGCTTAATCATGTGGT-3′ |
Metabion | N/A |
| Mouse Gfpt1 PCR cloning: EcoRI-Gfpt1 5′-ATGAATTCGTGAC CAACATCATGTGCGG-3′ |
Metabion | N/A |
| Mouse Gfpt1 PCR cloning: Gfpt1-STP-XhoI 5′-ATCTCGAGTTAC TCTACTGTTACAGATTTGGC-3′ |
Metabion | N/A |
| Gfpt1 PCR mutagenesis (removal of internal EcoRI site), forward primer 5′-CTCATTATTTTTATCAGAGCGCTGG-3′ |
Metabion | N/A |
| Gfpt1 PCR mutagenesis (removal of internal EcoRI site), reverse primer 5′-TTCATTGTTATTCATAATGGAATCATCA-3′ |
Metabion | N/A |
| Gfpt1 E328K PCR mutagenesis, forward primer 5′-TCTTCTGCATAAATGAACTAAAGTTG-3′ |
Metabion | N/A |
| Gfpt1 E328K PCR mutagenesis, reverse primer 5′-AAATTTTTGAGCAGCCAGAATCTG-3′ |
Metabion | N/A |
| Recombinant DNA | ||
| Wild-type and E328K mutant Mus musculus Gfpt1 cDNA PCR amplified and cloned into pBluescript and Sleeping Beauty donor plasmid | This paper | GenBank accession for Gfpt1 cDNA sequence NM_013528 |
| Software and Algorithms | ||
| GraphPad Prism 9 | GraphPad Software | www.graphpad.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Jukka Kallijärvi (jukka.kallijarvi@helsinki.fi).
Materials availability
The bacterial expression plasmid for OGT, and the GFPT1-overexpressing AML12 cells and the Sleeping Beauty transposon donor plasmid required to generate these cells are available upon request. Requests related to previously published non-commercial materials should be addressed to the corresponding authors of the publications cited.
Experimental model and study participant details
Mouse tissue samples
The laboratory animal center of the University of Helsinki maintained the wild-type mice of congenic C57BL/6JCrl background under the internal license of the research group (KEK22-018). The mice were housed in temperature-controlled (23°C) individually-ventilated cages with enriched environment under 12-h light/dark cycle and received chow (Teklad 2018, Harlan) ad libitum. Male and female mice of 1 month of age were euthanized by cervical dislocation and tissues immediately excised and placed in liquid N2 and stored at −80°C.
For comparison of UDP-HexNAc and UDP-GlcNAc levels, we utilized 23 samples from our other studies comprising liver extracts from 1-month old male and female mice of four different genotypes: wild-type, Bcs1lp.S78G heterozygotes and homozygotes, and Bcs1lp.S78G homozygotes with transgenic expression of Ciona intestinalis alternative oxidase.49 The genotype differences were irrelevant for this study, and the sole purpose for including these different genotypes was to maximize the number of data points and sufficient variation to allow a meaningful correlation analysis. The transgenic mice were bred under the permit ESAVI/16278/2020 by the State Provincial Office of Southern Finland.
GFPT1 knockout cells
The TU8988T GFPT1 knockout (clone B9 in ref. 38) and the parental cell lines were a kind gift from Prof. Costas A. Lyssiotis laboratory (University of Michigan, USA).
Cell culture
AML12 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) mixture F12 (DMEM-F12) with 5 mM glucose and supplemented with 10% fetal bovine serum, penicillin and streptomycin, 2 mM L-alanyl-L-glutamine, 10 μg/mL insulin, 5.5 μg/mL transferrin, and 5 ng/mL selenium. Other cell lines were maintained in DMEM with 5 mM glucose, 10% fetal bovine serum, penicillin and streptomycin, and 2 mM L-alanyl-L-glutamine. This media was supplemented with 10 mM GlcNAc to maintain the TU89888T GFPT1-knockout cells. All cells were cultured at 37°C in an atmosphere of 5% CO2 and 95% air.
For experiments in Figures 3 and 4, the cells were grown in T-25 cell culture flasks to ∼60% confluence using conventional culture conditions. After this, the medium composition was altered as stated in the figures and the figure legends. Unless an experimental variable, the glucose concentration in media was 5 mM. In the case of GFPT1 knock-out cells, the experiments were started at low cell density due to the expected proliferation of the cell during the experiment. To collect samples, the cells were washed once with PBS and detached with trypsin and EDTA (TrypLE, ThermoFisher Scientific) and counted. The cells were pelleted by centrifugation, resuspended in 60% MeOH, and stored at −80°C until continuing with the extraction (typically after 1–3 days).
Method details
Generation of GFPT1 overexpressing cell lines
The Mus musculus Gfpt1 cDNA was PCR-amplified (primers: EcoRI-Gfpt1 5′-ATGAATTCGTGACCAACATCATGTGCGG-3′ and Gfpt1-STP-XhoI 5′-ATCTCGAGTTACTCTACTGTTACAGATTTGGC-3′) and the PCR product ligated into pBluescript vector. Internal EcoRI site was removed by site-directed mutagenesis (primers: 5′-CTCATTATTTTTATCAGAGCGCTGG-3′ and 5′-TTCATTGTTATTCATAATGGAATCATCA-3′). The point mutation E328K was introduced by mutagenesis using the following primers: 5′-TCTTCTGCATAAATGAACTAAAGTTG-3′ and 5′-AAATTTTTGAGCAGCCAGAATCTG-3′). After sequencing, the wild type (WT) and E328K variants were subcloned into the Sleeping Beauty transposon donor plasmid ITR\CAG-MCS-IRES-Puro2A-Thy1.1\ITR. To generate stable cell line overexpressing GFPT1, AML12 cells were co-transfected with the Sleeping Beauty transposon donor plasmid carrying WT or mutant Gfpt1 and a plasmid encoding SB100X transposase (1:10 ratio) using FuGENE HD transfection reagent. Subsequently, the cells were subjected to puromycin selection (2 μg/ml). Resistant colonies were pooled and amplified. The expression of GFPT1 was confirmed by Western blot analyses using rabbit anti-GFPT1 antibody (Abcam, #ab125069, clone EPR4854).
Recombinant proteins
For initial assay development using the dot blotting format, we used commercial recombinant human OGT fragment (Cys323-Glu1041) (R&D systems, 8446-GT). Thereafter, we switched to in-house produced enzyme. A fragment of human OGT cDNA encoding amino acids 322–1041 was PCR-amplified from a human cDNA library (primers MunI-OGT 5′-ATCAATTGACCATGCTGTGTCCCACCCATGCAGA-3′ and OGT-STP-SalI 5′-ATATGTCGACTTATTCAACAGGCTTAATCATGTGGT-3′) and ligated into the EcoRI and XhoI sites of the bacterial expression vector pHAT2, in frame with an N-terminal hexahistidine tag. The plasmid was transformed into the BL21 E. coli strain. A single colony from the agar plate was inoculated into 5 mL LB medium containing 100 μg/mL ampicillin and cultured overnight. The bacteria were diluted to 100 mL LB and grown for 3–4 h at room temperature before addition of 0.5 mM IPTG to induce the OGT expression during overnight culture at room temperature. We tested two OGT purification protocols employing Ni-NTA Agarose (Qiagen, # 30210) or HisPur cobalt resin (Thermo Scientific, #89964) to capture the His-tagged protein. The latter approach yielded slightly purer product (Figure S5A) and higher enzymatic activity. This method is described here. The alternative approach, Ni-NTA-based purification, was performed essentially according to the manufacturer’s protocol. The bacteria were pelleted and suspended into 9 mL of lysis buffer comprising 0.5 mg/mL lysozyme, 25 U/ml benzonase, 1 mM MgCl2, 0.1% Triton X-100, 0.1 mM TCEP, protease inhibitor mix (Roche, #11873580001), and 0.5xPBS (5 mM Na-phosphate, 75 mM NaCl, pH 7.4). The bacterial cell walls were digested for 15 min at room temperature and 15 min on ice. Thereafter, the purification was continued while maintaining cold temperature (0-8°C) of the sample and buffers. The lysis was completed by three rounds of 10s probe sonication on ice with 10s cooling time (amplitude 20%, Branson Digital Sonifier 250). Remaining insoluble material was removed by centrifugation (5 min, 20 000 g at 4°C). The supernatant was supplemented to contain 10 mM imidazole, and the phosphate and NaCl concentrations were increased to 20 mM and 300 mM, respectively, by addition of 10xPBS. This solution was incubated for 1h at ∼7°C with 0.25 mL of HisPur Cobalt resin. The beads were allowed to settle to the bottom of a protein purification column and washed 5 times with 10 mL of washing buffer comprising 10 mM imidazole and 0.1% Triton X-100 in 2xPBS. The elution was performed with 150 mM imidazole in washing buffer supplemented with 0.1 mM TCEP. Five 0.25 mL elutes were collected and analyzed by SDS-PAGE (Figure S5). The elution fractions 2 to 5 were combined and dialyzed twice (∼16h and ∼3h at ∼7°C) against 200 mL of 50 mM Bis-Tris (Cl+) pH 7, 20% glycerol, and 0.1 mM TCEP. The yield was estimated by Bradford assay and BSA standards. The purified OGT was stored in 50% glycerol at −20°C.
The following recombinant proteins were purchased from commercial sources: alkaline phosphatase (Thermo Scientific #EF0651), Antarctic phosphatase (New England BioLabs, #M0289), and human casein kinase II (New England BioLabs, #P6010).
Preparation of GlcNAc-acceptor peptide-BSA complex
A peptide (NH2-KKKYPGGSTPVSSANMM-COOH) containing an O-GlcNAcylation site of human Casein kinase II subunit alpha (underlined sequence) was custom synthesized by Nordic BioSite. A mixture of 6 mM glutaraldehyde, 1 mg/mL peptide, and 1 mg/mL fatty-acid free BSA was incubated in phosphate-buffered saline (PBS) for 30 min to crosslink the peptide to BSA. The reaction was quenched by addition of 12 mM glycine. In our final protocol, the product was sonicated, clarified by centrifugation (3 min 22000g), and centrifuged through a 40 μm cell strainer to increase the uniformity of the coating and to decrease well-to-well variation. The solutions of peptide-BSA complex were stored at −20°C. For some initial experiments, we crosslinked the peptide using 20 mM glutaraldehyde (under the conditions above), quenched the reaction with 19 volumes of 10 mM Tris-HCl pH 7 and removed the uncrosslinked peptide with a Amicon Ultra-4 Centrifugal 10 kDa cut-off column. This approach, however, led to a partial precipitation of the product during centrifugation, and therefore we continued to use the former approach.
Preparation of tissue and cell extracts
Tissue pieces were disrupted in ice-cold 60% MeOH (typically 500 μL per 10 mg tissue) using microtube pestle homogenizer (10s) (Nippon Genetics, cat. no. NG010) or 20 strokes with roughened glass-to-glass potter homogenizer (for heart, skeletal muscle, and kidney) followed by sonication (12s, amplitude 10%, Branson Digital Sonifier 250) to complete the homogenization. Cell pellets were directly sonicated. Further precipitation of proteins was achieved by addition of 225 μL chloroform per 500 μL homogenate and centrifugation (3–6 min 18000 g at 4°C). The aqueous phase was collected and washed three times with ∼1.4 mL diethyl ether to remove MeOH. Any remaining layer of diethyl was evaporated by gentle ∼10s flow of N2 gas. The remaining traces of diethyl ether were evaporated at 65°C in centrifugal vacuum evaporator (SpeedVac Plus SC110A, Savant Instruments) for 5–6 min. The extract volume was estimated by weighing the extract and assuming density of 1 mg/μL. In some initial experiments, chloroform was used instead of diethyl ether to remove MeOH. For data from the TU8988T cells, the aqueous-methanol extracts after addition of chloroform were directly evaporated to dryness in miVac centrifugal evaporator (Genevac) without applied heat. The final extracts were stored at −80°C typically for 1–3 days before the UDP-GlcNAc measurements.
SDS-PAGE and western blotting
Precipitated proteins from the UDP-GlcNAc extraction were sedimented by addition of 800 μL MeOH and centrifugation (6 min 18000 g at 4°C). The protein pellets were dissolved in Laemmli buffer (2% SDS, 4% β-mercaptoethanol, 60 mM Tris-Cl pH 6.8, and 12% glycerol) with the help of sonication and 5 min incubation at 95°C. A modified Bradford reagent containing 2.5 mg/mL α-cyclodextrin to chelate the interfering SDS was employed to measure protein concentrations.50 The samples were run on Bio-Rad Criterion TGX gels followed by tank transfer onto PVDF membrane. Equal loading and transfer were verified by Coomassie G-250 staining. After complete destaining of Coomassie G-250, the membrane was blocked with 0.7% Na-caseinate pH 7.4–7.6 and 1% BSA and probed with the following primary antibodies: O-GlcNAc (clone RL2, BioLegend), OGT (ab96718, Abcam), OGA (HPA036141, Merck), and α-tubulin (clone DM1A, Cell Signaling Technology). Peroxidase-conjugated secondary antibodies (#7074 and #7076, Cell Signaling Technology) and enhanced chemiluminescence (ECL) were used for the detection. The ECL reagent51 comprised 0.75 mM luminol, 1 mM 4-(Imidazol-1-yl)phenol, 2 mM H2O2, and 0.05% Tween 20 in 10 mM Tris-Cl buffer pH 9. The luminescence was recorded with Bio-Rad ChemiDoc MP imager.
UDP-GlcNAc assay in dot blot format
The assay conditions were varied during the method development. Here, the final established assay is described. The O-GlcNAcylation reactions comprised: 10 μg/mL OGT, 25 U/ml alkaline phosphatase, 5 mM Mg-acetate, and 0.2 mg/mL OGT-substrate peptide-BSA complex (concentration based on BSA content), and 50 mM Bis-Tris pH 7.0 (adjusted with HCl). The reactions were carried out in 10 μL volume for 2h at room temperature (21-23°C). The sample volume comprised 2 μL of the reaction mixture. The reactions were stopped by dropping the temperature to 0°C. Bio-Rad microfiltration blotting device was employed to capture the OGT-substrate peptide-BSA complex onto 0.2 μm PVDF membrane (Amersham Hybond P). After completion of the dot blot, the membrane was completely dried, soaked in MeOH, rehydrated, and blocked with 0.7% Na-Caseinate pH 7.4–7.6, 1% BSA for 1h. O-GlcNAcylated residues were detected with the mouse monoclonal RL2 antibody (0.5 μg/mL, 2h), peroxidase-conjugated secondary antibody, and enhanced chemiluminescence.
UDP-GlcNAc assay in microplate format
The assay parameters were varied and optimized throughout this study. The final established assay conditions are described here. The wells of Nunc MaxiSorp 384 plate were coated with 20 μL of 10 μg/mL OGT-substrate peptide-BSA complex (100 ng BSA, 100 ng peptide) in PBS for ∼16 h at +4°C. The wells were washed twice with 115 μL of TBS containing 0.05% tween 20 (TBST) and once with 50 mM Bis-Tris buffer pH 7.0. The plate was placed on ice and 16 μL of a reagent mix was quickly added into the wells followed by 4 μL of standard or diluted extract. The reagent mix comprised 18.75 μg/mL OGT, 31.25 U/ml alkaline phosphatase, 6.25 mM Mg-acetate, 0.375 mg/mL fatty-acid free BSA, and 62.5 mM Bis-Tris (HCl) pH 7.0. The reactions were carried out for 2-3h at room temperature (21-23°C). The wells were washed twice with TBST and once with TBS. Then, the wells were incubated with 20 μL of RL2 antibody (0.5 μg/mL) for 1-2h. Following 5 washes with TBST and one wash with TBS, 20 μL of peroxidase-conjugated secondary antibody (1:3000, #7076, Cell Signaling Technology) was added into each well and incubated for 0.5-1h. Both antibodies were diluted in 1% BSA in TBST (0.2% Tween 20). After repetition of the washing steps, 12.5 μM Amplex UltraRed (Invitrogen, #A36006) and 1 mM H2O2 were utilized as the peroxidase substrates in 100 mM potassium phosphate buffer pH 6.7. The chemifluorescence was developed for 45 min in dark and the end-point fluorescence read upon 530 nm excitation and 590 nm emission with BioTek Synergy H1 plate reader. To prevent evaporation during the incubation steps the plates were sealed with an adhesive membrane.
UPLC-MS-based quantification of UDP-HexNAc
To approximately 15 mg of snap-frozen liver samples, 600 μL of ice-cold chloroform-methanol (2:1) solution was added. The tissue pieces were ground with a ball mill and subjected to three 10-min ultrasonic bath treatments on ice and three freeze-thaw cycles (−196°C–4°C). Samples were vortexed in cold room (4°) for 30 min prior to addition of 400 μL milli-Q water. After addition of water, samples were further vortexed for 30 min and centrifuged to induce phase separation. Upper water-methanol phase was transferred to a new Eppendorf tube and evaporated to dryness in vacuum concentrator (Genevac, miVac Duo). Samples were reconstituted in 100 μL water and analyzed with UPLC-QTRAP/MS (ABSciex).
Chromatographic separation was performed in Waters Premier BEH C18 AX column (150 × 2.1mm, ø 1.7μm) at 40°C. Elution solvents were 10 mM ammonium acetate in water, pH 9.0 (A) and acetonitrile (ACN) (B) with flow rate of 0.3 mL min−1. Linear gradient started with 98% A and decreased to 30% in 3 min, then to 10% in 1 min, back to 98% in 4.01 min time, and stabilized for 1 min, with 5 min total runtime. Injection volume was 1 μL. UDP-HexNAc was analyzed with UPLC-6500 + QTRAP/MS (ABSciex) in negative ion mode. Retention time was 1.32 min. Multiple Reaction Monitoring (MRM) method with two transitions, quantitative and qualitative were performed: quantitative 606 → 385, and qualitative 606 → 159.
After the UPLC-MS runs, the extracts were evaporated to dryness, and stored (∼1.5 years) at −80°C, before measurement of UDP-GlcNAc with the enzymatic method.
Quantification and statistical analysis
The signal-to-noise ratio was defined as background-subtracted fluorescence divided by the standard deviation of the background (n > 6). LLOQ and LLOD were estimated empirically (variation and standard curve fit) and mathematically based on signal-to-noise cut-off of 10 and 2, respectively. Signal-to-background was defined as signal divided by the background (0 nM UDP-GlcNAc). The bar graphs for data from biological replicates represent mean and standard deviation (SD). Mean and standard error of mean (SEM) are shown for technical replicates. GraphPad Prism (versions 9.3.1–9.5.0) was utilized for non-linear curve fitting and other statistical analyses. Differences between the groups were assessed with two-tailed t-test or one-way ANOVA followed by appropriate multiple comparisons tests as stated in the figure legends.
Acknowledgments
We thank Prof. Costas Lyssiotis and Dr. Lin Lin (University of Michigan, USA) for providing the GFPT1 knockout cell lines and for valuable advice regarding their culture. The Methanococcus maripaludis UDP-GlcNAc dehydrogenase bacterial expression construct was a kind gift by Dr. David E. Graham (Oak Ridge National Laboratory, USA). Part of the work was carried out with the support of HiLIFE Laboratory Animal Center Core Facility, University of Helsinki, Finland. We acknowledge funding from Samfundet Folkhälsan (V.F. and J.K.), the Jane and Aatos Erkko Foundation (J.K.), the Foundation for Pediatric Research (V.F.), Finska Läkaresällskapet (V.F.), the Liv och Hälsa Foundation (V.F.), the Magnus Ehrnrooth Foundation (M.S.), and the Alfred Kordelin Foundation (J.P.). The graphical illustrations were created with the help of BioRender.com.
Author contributions
J.P. invented the assay concept and wrote the first manuscript draft. J.K. was responsible for the cloning of the OGT construct. J.K., M.S., and J.P. took part in the production of the recombinant OGT. J.P., M.S., D.U., and R.B. performed the UDP-GlcNAc measurements. M.S., D.U., and R.B. were responsible for the cell culture work. D.U. cloned the GFPT1 plasmids and generated the GFPT1-overexpressing cell lines. N.S. performed the UPLC-MS measurements. All authors critically read and commented on the manuscript, and J.P. and J.K. revised it accordingly. J.P. and J.K. supervised the study.
Declaration of interests
The authors declare no competing interests.
Published: June 28, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2023.100518.
Contributor Information
Jukka Kallijärvi, Email: jukka.kallijarvi@helsinki.fi.
Janne Purhonen, Email: janne.purhonen@helsinki.fi.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Chatham J.C., Zhang J., Wende A.R. Role of O-linked N-acetylglucosamine protein modification in cellular (patho)physiology. Physiol. Rev. 2021;101:427–493. doi: 10.1152/physrev.00043.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiaradonna F., Ricciardiello F., Palorini R. The nutrient-sensing hexosamine biosynthetic pathway as the hub of cancer metabolic rewiring. Cells. 2018;7 doi: 10.3390/cells7060053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanover J.A., Chen W., Bond M.R. O-GlcNAc in cancer: an Oncometabolism-fueled vicious cycle. J. Bioenerg. Biomembr. 2018;50:155–173. doi: 10.1007/s10863-018-9751-2. [DOI] [PubMed] [Google Scholar]
- 4.Bond M.R., Hanover J.A. O-GlcNAc cycling: a link between metabolism and chronic disease. Annu. Rev. Nutr. 2013;33:205–229. doi: 10.1146/annurev-nutr-071812-161240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang X., Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2017;18:452–465. doi: 10.1038/nrm.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buse M.G., Robinson K.A., Gettys T.W., McMahon E.G., Gulve E.A. Increased activity of the hexosamine synthesis pathway in muscles of insulin-resistant ob/ob mice. Am. J. Physiol. 1997;272:E1080–E1088. doi: 10.1152/ajpendo.1997.272.6.E1080. [DOI] [PubMed] [Google Scholar]
- 7.Yki-Järvinen H., Virkamäki A., Daniels M.C., McClain D., Gottschalk W.K. Insulin and glucosamine infusions increase O-linked N-acetyl-glucosamine in skeletal muscle proteins in vivo. Metabolism. 1998;47:449–455. doi: 10.1016/S0026-0495(98)90058-0. [DOI] [PubMed] [Google Scholar]
- 8.Robinson K.A., Willi S.M., Bingel S., Buse M.G. Decreased hexosamine biosynthesis in GH-deficient dwarf rat muscle. Reversal with GH, but not IGF-I, therapy. Am. J. Physiol. 1999;276:E435–E442. doi: 10.1152/ajpendo.1999.276.3.E435. [DOI] [PubMed] [Google Scholar]
- 9.Hawkins M., Angelov I., Liu R., Barzilai N., Rossetti L. The tissue concentration of UDP-N-acetylglucosamine modulates the stimulatory effect of insulin on skeletal muscle glucose uptake. J. Biol. Chem. 1997;272:4889–4895. doi: 10.1074/jbc.272.8.4889. [DOI] [PubMed] [Google Scholar]
- 10.Lehmann R., Huber M., Beck A., Schindera T., Rinkler T., Houdali B., Weigert C., Häring H.U., Voelter W., Schleicher E.D. Simultaneous, quantitative analysis of UDP-N-acetylglucosamine, UDP-N-acetylgalactosamine, UDP-glucose and UDP-galactose in human peripheral blood cells, muscle biopsies and cultured mesangial cells by capillary zone electrophoresis. Electrophoresis. 2000;21:3010–3015. doi: 10.1002/1522-2683(20000801)21:14<3010::AID-ELPS3010>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 11.Haltiwanger R.S., Blomberg M.A., Hart G.W. Glycosylation of nuclear and cytoplasmic proteins. Purification and characterization of a uridine diphospho-N-acetylglucosamine:polypeptide beta-N-acetylglucosaminyltransferase. J. Biol. Chem. 1992;267:9005–9013. doi: 10.1016/S0021-9258(19)50380-5. [DOI] [PubMed] [Google Scholar]
- 12.Kreppel L.K., Hart G.W. Regulation of a cytosolic and nuclear O-GlcNAc transferase: role of the tetratricopeptide repeats. J. Biol. Chem. 1999;274:32015–32022. doi: 10.1074/jbc.274.45.32015. [DOI] [PubMed] [Google Scholar]
- 13.Iyer S.P.N., Hart G.W. Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J. Biol. Chem. 2003;278:24608–24616. doi: 10.1074/jbc.M300036200. [DOI] [PubMed] [Google Scholar]
- 14.Perez M., Hirschberg C.B. Translocation of UDP-N-acetylglucosamine into vesicles derived from rat liver rough endoplasmic reticulum and Golgi apparatus. J. Biol. Chem. 1985;260:4671–4678. doi: 10.1016/S0021-9258(18)89122-0. [DOI] [PubMed] [Google Scholar]
- 15.Weckbecker G., Keppler D.O. Separation and analysis of 4′-epimeric UDP-sugars by borate high-performance liquid chromatography. Anal. Biochem. 1983;132:405–412. doi: 10.1016/0003-2697(83)90027-1. [DOI] [PubMed] [Google Scholar]
- 16.Hull S.R., Montgomery R. Separation and analysis of 4′-epimeric UDP-sugars, nucleotides, and sugar phosphates by anion-exchange high-performance liquid chromatography with conductimetric detection. Anal. Biochem. 1994;222:49–54. doi: 10.1006/abio.1994.1452. [DOI] [PubMed] [Google Scholar]
- 17.Tomiya N., Ailor E., Lawrence S.M., Betenbaugh M.J., Lee Y.C. Determination of nucleotides and sugar nucleotides involved in protein glycosylation by high-performance anion-exchange chromatography: sugar nucleotide contents in cultured insect cells and mammalian cells. Anal. Biochem. 2001;293:129–137. doi: 10.1006/abio.2001.5091. [DOI] [PubMed] [Google Scholar]
- 18.Namboori S.C., Graham D.E. Enzymatic analysis of uridine diphosphate N-acetyl-d -glucosamine. Anal. Biochem. 2008;381:94–100. doi: 10.1016/j.ab.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi J., Wang R., Zeng Y., Yu W., Gu Y. New ELISA-based method for the detection of O-GlcNAc transferase activity in vitro. Prep. Biochem. Biotechnol. 2017;47:699–702. doi: 10.1080/10826068.2017.1303614. [DOI] [PubMed] [Google Scholar]
- 20.Alteen M.G., Gros C., Meek R.W., Cardoso D.A., Busmann J.A., Sangouard G., Deen M.C., Tan H.-Y., Shen D.L., Russell C.C., et al. A direct fluorescent activity assay for glycosyltransferases enables convenient high-throughput screening: application to O-GlcNAc transferase. Angew. Chem., Int. Ed. 2020;59:9601–9609. doi: 10.1002/anie.202000621. [DOI] [PubMed] [Google Scholar]
- 21.Marshall S., Duong T., Orbus R.J., Rumberger J.M., Okuyama R. Measurement of UDP-N-acetylglucosaminyl transferase (OGT) in brain cytosol and characterization of anti-OGT antibodies. Anal. Biochem. 2003;314:169–179. doi: 10.1016/S0003-2697(02)00686-3. [DOI] [PubMed] [Google Scholar]
- 22.Holt G.D., Snow C.M., Senior A., Haltiwanger R.S., Gerace L., Hart G.W. Nuclear pore complex glycoproteins contain cytoplasmically disposed O-linked N-acetylglucosamine. J. Cell Biol. 1987;104:1157–1164. doi: 10.1083/jcb.104.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson R., Creavin A., O’Connell M., O’Connor B., Clarke P. Optimization of the enzyme-linked lectin assay for enhanced glycoprotein and glycoconjugate analysis. Anal. Biochem. 2011;413:114–122. doi: 10.1016/j.ab.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Haltiwanger R.S., Holt G.D., Hart G.W. Enzymatic addition of O-GlcNAc to nuclear and cytoplasmic proteins. Identification of a uridine diphospho-N-acetylglucosamine:peptide beta-N-acetylglucosaminyltransferase. J. Biol. Chem. 1990;265:2563–2568. doi: 10.1016/S0021-9258(19)39838-2. [DOI] [PubMed] [Google Scholar]
- 25.Mohammad K., Esen A. A blocking agent and a blocking step are not needed in ELISA, immunostaining dot-blots and Western blots. J. Immunol. Methods. 1989;117:141–145. doi: 10.1016/0022-1759(89)90129-4. [DOI] [PubMed] [Google Scholar]
- 26.Ritter J.B., Genzel Y., Reichl U. Simultaneous extraction of several metabolites of energy metabolism and related substances in mammalian cells: optimization using experimental design. Anal. Biochem. 2008;373:349–369. doi: 10.1016/j.ab.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 27.Fülöp N., Feng W., Xing D., He K., Nőt L.G., Brocks C.A., Marchase R.B., Miller A.P., Chatham J.C. Aging leads to increased levels of protein O-linked N-acetylglucosamine in heart, aorta, brain and skeletal muscle in Brown-Norway rats. Biogerontology. 2008;9:139. doi: 10.1007/s10522-007-9123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olson A.K., Bouchard B., Zhu W.Z., Chatham J.C., Des Rosiers C. First characterization of glucose flux through the hexosamine biosynthesis pathway (HBP) in ex vivo mouse heart. J. Biol. Chem. 2020;295:2018–2033. doi: 10.1074/jbc.RA119.010565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor R.P., Parker G.J., Hazel M.W., Soesanto Y., Fuller W., Yazzie M.J., McClain D.A. Glucose deprivation stimulates O-GlcNAc modification of proteins through up-regulation of O-linked N-acetylglucosaminyltransferase. J. Biol. Chem. 2008;283:6050–6057. doi: 10.1074/jbc.M707328200. [DOI] [PubMed] [Google Scholar]
- 30.Taylor R.P., Geisler T.S., Chambers J.H., McClain D.A. Up-regulation of O-GlcNAc transferase with glucose deprivation in HepG2 cells is mediated by decreased hexosamine pathway flux. J. Biol. Chem. 2009;284:3425–3432. doi: 10.1074/jbc.M803198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosch R.R., Pouwels M.-J.J.M., Span P.N., Olthaar A.J., Tack C.J., Hermus A.R.M.M., Sweep C.G.J., Sweep C.G.J. Hexosamines are unlikely to function as a nutrient-sensor in 3T3-L1 adipocytes. Endocrine. 2004;23:17–24. doi: 10.1385/ENDO:23:1:17. [DOI] [PubMed] [Google Scholar]
- 32.Wu J.C., Merlino G., Fausto N. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc. Natl. Acad. Sci. USA. 1994;91:674–678. doi: 10.1073/pnas.91.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du X.-L., Edelstein D., Rossetti L., Fantus I.G., Goldberg H., Ziyadeh F., Wu J., Brownlee M. Hyperglycemia-induced mitochondrial superoxide overproduction activates the hexosamine pathway and induces plasminogen activator inhibitor-1 expression by increasing Sp1 glycosylation. Proc. Natl. Acad. Sci. USA. 2000;97:12222–12226. doi: 10.1073/pnas.97.22.12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moiz B., Garcia J., Basehore S., Sun A., Li A., Padmanabhan S., Albus K., Jang C., Sriram G., Clyne A.M. 13C metabolic flux analysis indicates endothelial cells attenuate metabolic perturbations by modulating TCA activity. Metabolites. 2021;11:226. doi: 10.3390/metabo11040226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basehore S.E., Bohlman S., Weber C., Swaminathan S., Zhang Y., Jang C., Arany Z., Clyne A.M. Laminar flow on endothelial cells suppresses eNOS O-GlcNAcylation to promote eNOS activity. Circ. Res. 2021;129:1054–1066. doi: 10.1161/CIRCRESAHA.121.318982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z., Tan E.P., VandenHull N.J., Peterson K.R., Slawson C. O-GlcNAcase expression is sensitive to changes in O-GlcNAc homeostasis. Front. Endocrinol. 2014;5:206. doi: 10.3389/fendo.2014.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hugo S.E., Schlegel A. A genetic model to study increased hexosamine biosynthetic flux. Endocrinology. 2017;158:2420–2426. doi: 10.1210/en.2017-00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim P.K., Halbrook C.J., Kerk S.A., Radyk M., Wisner S., Kremer D.M., Sajjakulnukit P., Andren A., Hou S.W., Trivedi A., et al. Hyaluronic acid fuels pancreatic cancer cell growth. Elife. 2021;10 doi: 10.7554/eLife.62645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zachara N.E., O’Donnell N., Cheung W.D., Mercer J.J., Marth J.D., Hart G.W. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress: a survival response of mammalian cells. J. Biol. Chem. 2004;279:30133–30142. doi: 10.1074/jbc.M403773200. [DOI] [PubMed] [Google Scholar]
- 40.Kazemi Z., Chang H., Haserodt S., McKen C., Zachara N.E. O-linked β-N-acetylglucosamine (O-GlcNAc) regulates stress-induced heat shock protein expression in a GSK-3β-dependent manner. J. Biol. Chem. 2010;285:39096–39107. doi: 10.1074/jbc.M110.131102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian K., Wang S., Fu M., Zhou J., Singh J.P., Li M.-D., Yang Y., Zhang K., Wu J., Nie Y., et al. Transcriptional regulation of O-GlcNAc homeostasis is disrupted in pancreatic cancer. J. Biol. Chem. 2018;293:13989–14000. doi: 10.1074/jbc.RA118.004709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gloster T.M., Zandberg W.F., Heinonen J.E., Shen D.L., Deng L., Vocadlo D.J. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat. Chem. Biol. 2011;7:174–181. doi: 10.1038/nchembio.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheung W.D., Hart G.W. AMP-activated protein kinase and p38 MAPK activate O-GlcNAcylation of neuronal proteins during glucose deprivation. J. Biol. Chem. 2008;283:13009–13020. doi: 10.1074/jbc.M801222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang J.G., Park S.Y., Ji S., Jang I., Park S., Kim H.S., Kim S.-M., Yook J.I., Park Y.-I., Roth J., Cho J.W. O-GlcNAc protein modification in cancer cells increases in response to glucose deprivation through glycogen degradation. J. Biol. Chem. 2009;284:34777–34784. doi: 10.1074/jbc.M109.026351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou L., Zhu-Mauldin X., Marchase R.B., Paterson A.J., Liu J., Yang Q., Chatham J.C. Glucose deprivation-induced increase in protein O-GlcNAcylation in cardiomyocytes is calcium-dependent. J. Biol. Chem. 2012;287:34419–34431. doi: 10.1074/jbc.M112.393207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paneque A., Fortus H., Zheng J., Werlen G., Jacinto E. The hexosamine biosynthesis pathway: regulation and function. Genes. 2023;14:933. doi: 10.3390/genes14040933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma J., Wu C., Hart G.W. Analytical and biochemical perspectives of protein O-GlcNAcylation. Chem. Rev. 2021;121:1513–1581. doi: 10.1021/acs.chemrev.0c00884. [DOI] [PubMed] [Google Scholar]
- 48.Ma X., Liu P., Yan H., Sun H., Liu X., Zhou F., Li L., Chen Y., Muthana M.M., Chen X., et al. Substrate specificity provides insights into the sugar donor recognition mechanism of O-GlcNAc transferase (OGT) PLoS One. 2013;8 doi: 10.1371/journal.pone.0063452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rajendran J., Purhonen J., Tegelberg S., Smolander O.-P., Mörgelin M., Rozman J., Gailus-Durner V., Fuchs H., Hrabe de Angelis M., Auvinen P., et al. Alternative oxidase-mediated respiration prevents lethal mitochondrial cardiomyopathy. EMBO Mol. Med. 2019;11 doi: 10.15252/emmm.201809456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rabilloud T. Optimization of the cydex blue assay: a one-step colorimetric protein assay using cyclodextrins and compatible with detergents and reducers. PLoS One. 2018;13 doi: 10.1371/journal.pone.0195755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang L., Jin M., Du P., Chen G., Zhang C., Wang J., Jin F., Shao H., She Y., Wang S., et al. Study on enhancement principle and stabilization for the Luminol-H2O2-HRP chemiluminescence system. PLoS One. 2015;10 doi: 10.1371/journal.pone.0131193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.