Fig. 5. Identification of genes associated with aggressive OPSCC.
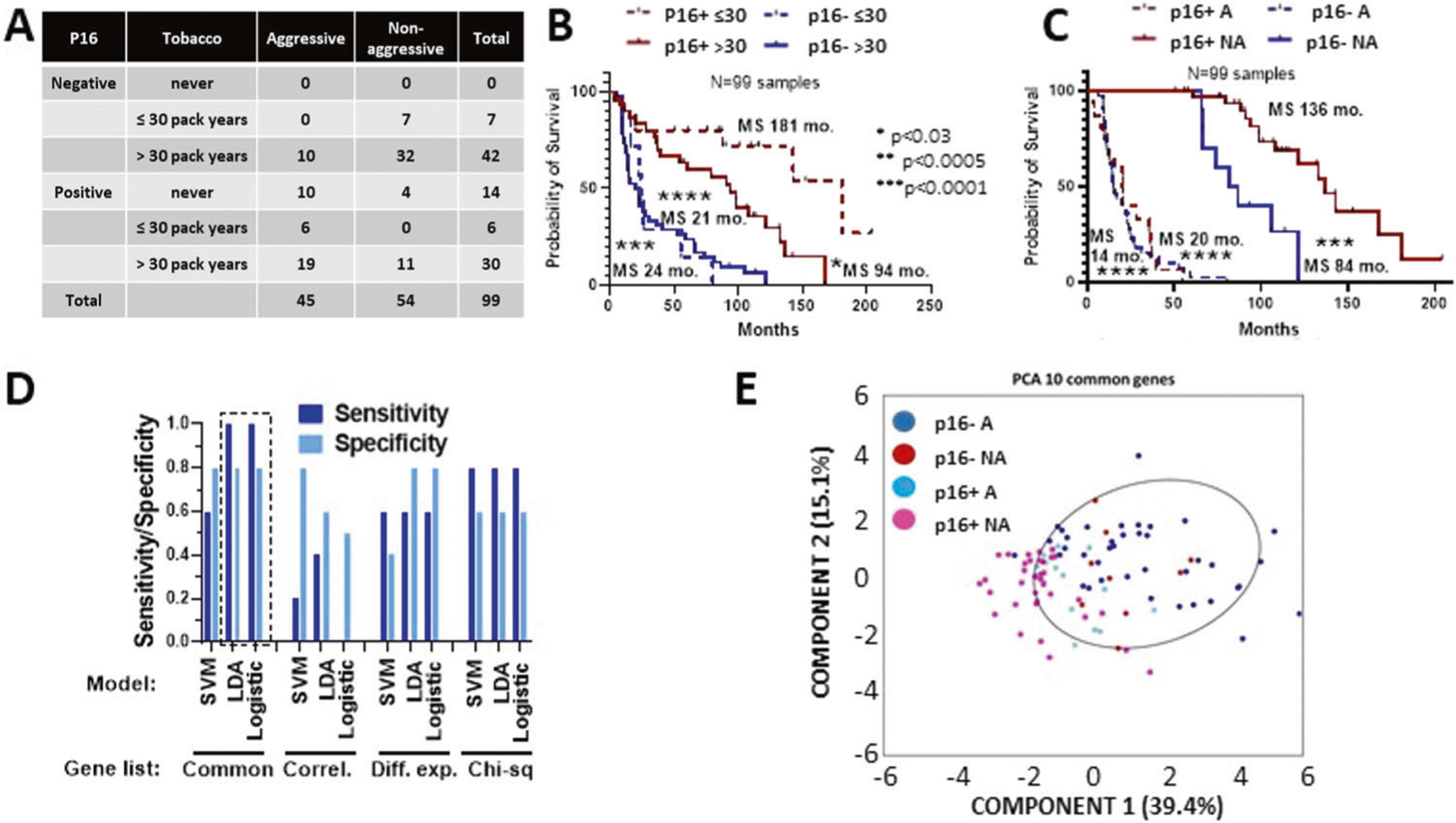
A Relevant clinical characteristics of cohort chosen for focused transcriptional gene expression analysis. Aggressive (A) disease was defined as associated with death or distant metastasis within 5 years; non-aggressive (NA) disease was defined as no death or distant metastasis within 5 years. B Survival of the RNA cohort stratified by tobacco exposure and p16 status. C Survival of RNA cohort stratified by aggressive (A) vs non-aggressive (NA) classification and p16 status. D Validation results from machine learning models using different gene selection lists: Common= 10 genes in common between top 20 ranking differentially expressed and most correlated genes; Correl = top 10 most correlated genes with survival time; Diff. exp. = top 10 most significantly different genes between aggressive and non-aggressive tumors; Chi-sq = top 10 genes using univariate ranking function in Matlab. E Principal component analysis (PCA) using RNA expression from the 10 genes on the common list demonstrating the genes could separate A p16+ and p16-neg tumors from NA p16+ OPSCC tumors.
