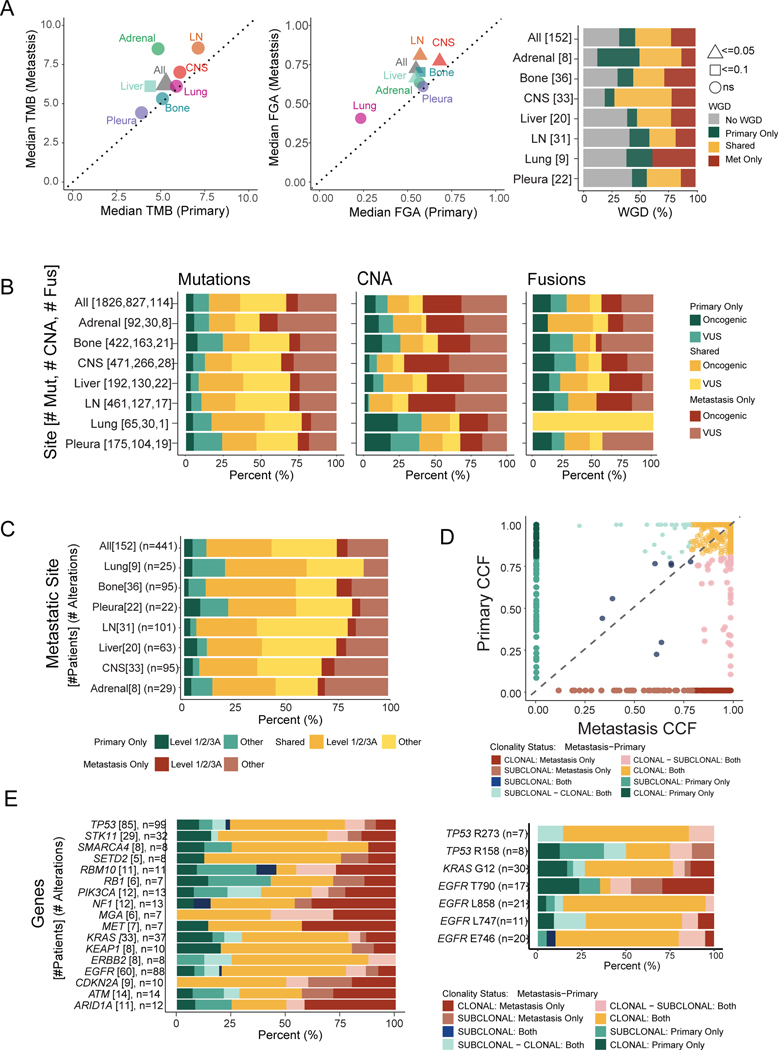
Figure 5. Genomic comparisons between patient-matched primary tumors and metastases.
(A) Median tumor mutational burden (TMB) and fraction of genome altered (FGA) between matched primary tumors and metastases. Statistical analysis: Wilcoxon rank-sum test. p-values as indicated. Right: Proportion of samples with whole-genome duplication (WGD) private or shared between primary and metastatic samples. (B) Distribution of shared and private alterations stratified by alteration types across sites. (C) Actionability of alterations private or shared between primary and metastatic samples across sites. (D) Cancer cell fraction (CCF) of shared mutations between primary and metastatic samples. CCF <0.8 is considered subclonal. (E) Proportion of shared and private mutations, stratified by clonality status for each gene (left) and specific amino acid changes (right). Clonality status nomenclature describes mutation clonality first in metastasis and then primary sample. CNA, copy number alteration; CNS, central nervous system; Fus, fusion; LN, lymph node; Met, metastasis; Mut, mutation; VUS, variant of unknown significance.
See also Figure S8.