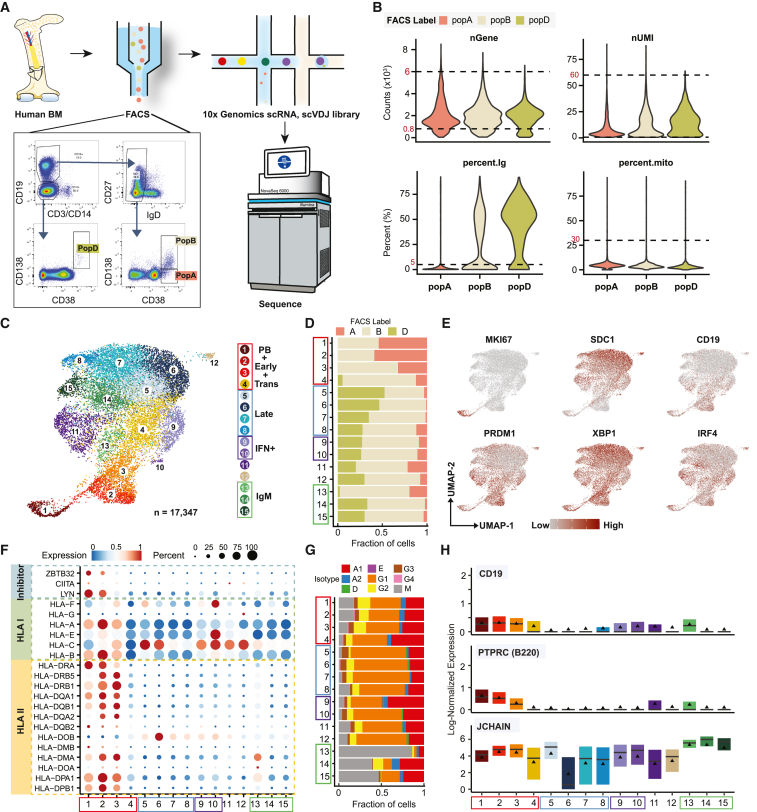
Figure 1.
Single-cell transcriptomic profiling of bone marrow PCs
(A) Schematic of single-cell RNA profiling for human BMPCs.
(B) Criterion for removing bad-quality cells. Dashed lines show the cutoffs that are labeled in red.
(C) scRNA-seq cell clusters of combined data from five healthy BMs and visualized in UMAP colored by cell types. Red and blue boxes highlight the early and late stage of BMPC maturation, respectively. The purple box highlights the path toward IFN-response PC subgroups and the green box highlights IgM-dominant cell populations.
(D) The fraction of cells from fluorescence-activated cell sorting (FACS)-sorted cell population in each cell subgroup identified in (C).
(E) Key PC-associated master gene expression. The redder the dot, the higher the log-normalized gene expression.
(F) Dot plot for expression of human MHC class I, II, and inhibitors of class II genes. Colors represent minimum-maximum normalized mean expression of marker genes in each cell group, and sizes indicate the proportion of cells expressing marker genes.
(G) The fraction of isotypes identified by scVDJ-seq data in each cell subgroup identified in (C).
(H) Boxplots showing the expression levels of the indicated genes. The solid triangle represents the average value.